Phosphoketolases for improved production of acetyl coenzyme a-derived metabolites, isoprene, isoprenoid precursors, and isoprenoids
一种类异戊二烯、异戊二烯合酶的技术,应用在生物化学设备和方法、微生物的测定/检验、酶等方向,能够解决损耗、生产收率降低等问题
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
example 1
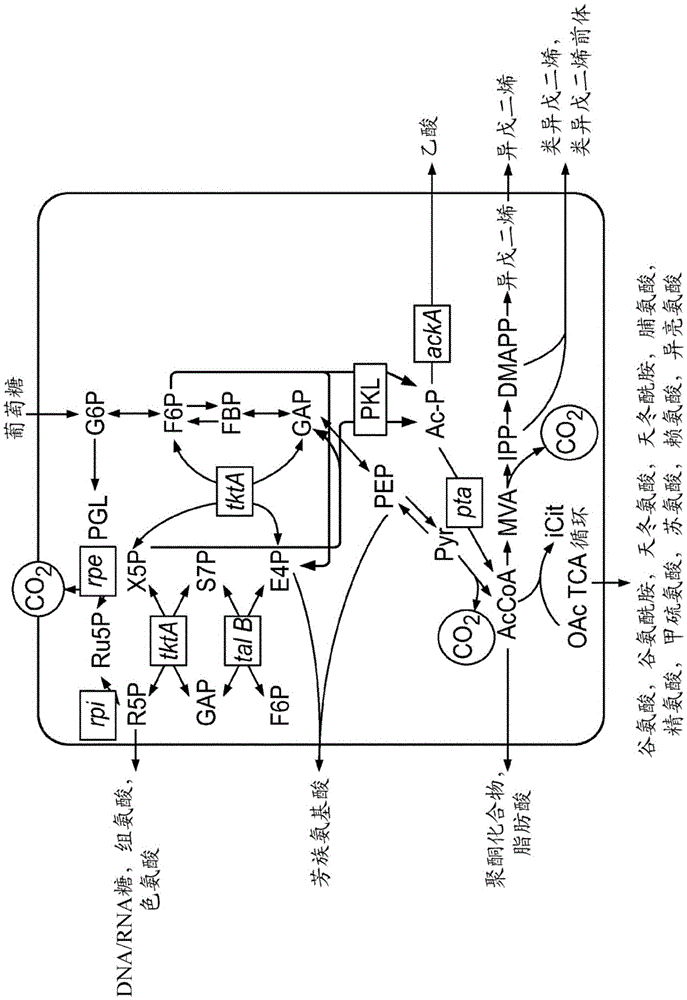
[0540] Example 1: Identification of Phosphoketolases
[0541] To identify phosphoketolases useful for improved production of acetyl-CoA-derived (acetyl-CoA-derived) metabolites, isoprenes, isoprenoid precursors, and isoprenoids in recombinant cells, in The CDART program in the NCBI network is used to select all gene products that are consistent with the known phosphoketolase domain structure (GeerL et al. (2002), "CDART: proteinhomology by domain architecture.", GenomeRes.12 (10) 1619- twenty three). Sequences were further refined by selecting RefSeq sequences from the initial domain construction search. Next, the sequences were divided into 22 different groups based on sequence similarity (Clustering by Passing Messages Between Data Points. Brendan J. Frey and Delbert Dueck, University of Toronto Science 315, 972-976, February 2007). Briefly, amino acid sequences were multiple-aligned using ClustalW. Pairwise percent identities (PIDs) were calculated. This is operationa...
example 2
[0556] Example 2: Identification of Phosphoketolases in Bacterial Genomes Deleting Phosphofructokinase
[0557] Bacterial genomes were searched for annotated phosphoketolase (PKL) but not phosphofructokinase (PFK), a key enzyme in carbon flux through glycolysis. Several organisms that fit these criteria were selected for high activity and high yield of glucose to isoprene for a specific PKL from this list of five PKLs from the plant Burkholderiaphytofirmans PsJN (SEQ ID NO: 47), Lactobacillus buchneri NRRLB-30929 (SEQ ID NO: 48), Bifidobacterium gallicum DSM20093 (SEQ ID NO: 49), Bifidobacterium dentium Bd1 (SEQ ID NO: 50 ) and Bifidobacterium bifidum IPLA20015 (SEQ ID NO: 51). Since most PKLs from the full repertoire of organisms have not yet been characterized, the selection of five PKLs was made based on sequence differences and the best environmental evidence of hyperactivity available from the literature. PKL from Bifidobacterium dentium shows a pH optimum of 7 (Sgorb...
example 3
[0558] Example 3: Cloning of identified phosphoketolases
[0559] PKL obtained from Bifidobacterium longum subsp. infantis, Enterococcus gallinarum and Clostridium acetobutylicum were each assayed for their enzymatic activity. Bifidobacterium longum subsp. infantis PKL has a Km of 5.7±1.16mM, 4.56±0.2sec -1 kcat, and 0.79±0.2mM -1 sec -1 kcat / Km, Enterococcus gallinarum (Enterococcus gallinarum) PKL has a Km of 10.4±1.03mM, 1.35±0.04sec -1 kcat, and 0.13±0.1mM -1 sec -1 kcat / Km, and Clostridium acetobutylicum (Clostridium acetobutylicum) PKL was found to have a Km of 10.3±0.67mM, 2.18±0.05sec -1 kcat, and 0.21±0.06mM -1 sec -1 kcat / Km. Constructs encoding Bifidobacterium longum subsp. infantis, Enterococcus gallinarum, or Clostridium acetobutylicum PKL were used as controls to screen candidate PKL enzymes for in vivo and in vitro activity.
[0560]The amino acid sequence (SEQ ID NO: 93) of Enterococcus gallinarum PKL was obtained from GenBank and processed in GeneAr...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com