Method for detecting 5-methylcystein in RNA and kit thereof
A technology of hydroxymethylcytosine and detection kits, applied in the field of biochemistry, can solve the problems of high cost, cumbersome operation, and long time consumption, and achieve the effects of easy operation, simple sample preparation, and high sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
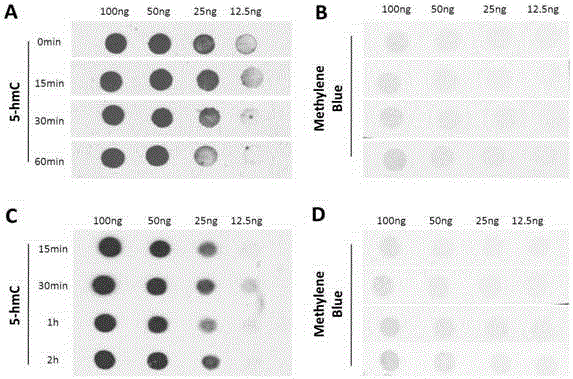
[0075] Example 1 RNA 5hmC detection method
[0076] 1. Extract total RNA and dilute to a concentration of 100ng / μL.
[0077] 2. Add an equal volume of 2M NaOH solution and react at 4°C for 30 minutes.
[0078] 3. Take a 4×8 nitrocellulose membrane, drop 2 μL of samples onto the membrane sequentially, and place it at room temperature for 15 minutes.
[0079] 4. Place the film in an oven at 80°C and bake for 30 minutes.
[0080] 5. After taking it out from 80°C, block it with 5% milk, react at room temperature for 1 hour, and then wash it with PBST solution 3 times, 10 minutes each time.
[0081] 6. Incubate the NC membrane with the 5hmC antibody diluted in PBS solution, react overnight at 4°C, and then wash with PBST solution 3 times, 10 minutes each time.
[0082] 7. Incubate the NC membrane with a specific secondary antibody diluted with 5% milk, react at room temperature for 1 hour, and then wash with PBST solution 3 times, 10 minutes each time.
[0083] 8. After develop...
Embodiment 2
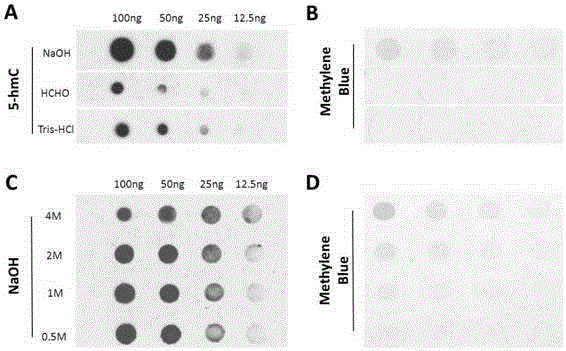
[0086] Example 2 Effects of Different Denaturing Agents and Concentrations on RNA 5hmC Detection
[0087] (1) The influence of different denaturing reagents on the detection effect of RNA 5hmC
[0088] Denaturing reagent type setting: NaOH, formaldehyde, Tris-HCl;
[0089] The test method is:
[0090] 1. Extract total RNA and dilute to a concentration of 100ng / μL.
[0091] 2. Add an equal volume of denaturing reagent solution (NaOH, formaldehyde or Tris-HCl) with a concentration of 2M, and react at 4°C for 30 minutes.
[0092] 3. Take a 4×8 nitrocellulose membrane, drop 2 μL of samples onto the membrane sequentially, and place it at room temperature for 15 minutes.
[0093] 4. Place the film in an oven at 80°C and bake for 30 minutes.
[0094] 5. After taking it out from 80°C, block it with 5% milk, react at room temperature for 1 hour, and then wash it with PBST solution 3 times, 10 minutes each time.
[0095] 6. Incubate the NC membrane with the 5hmC antibody diluted in...
Embodiment 3
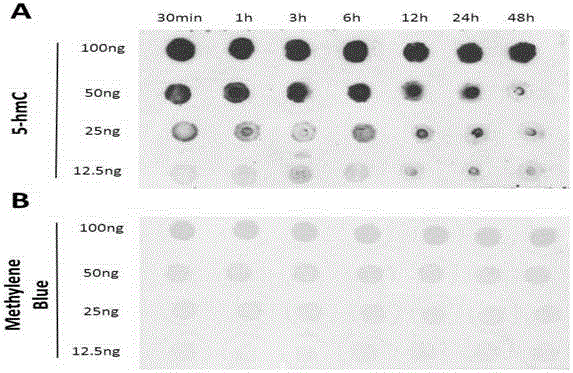
[0115] Example 3 Effects of different denaturation times on the detection effect of RNA 5hmC
[0116] Denaturation time setting: 30min, 1h, 3h, 6h, 12h, 24h, 48h;
[0117] The test method is:
[0118] 1. Extract total RNA and dilute to a concentration of 100ng / μL.
[0119] 2. Add an equal volume of NaOH with a concentration of 2M, and react at 4°C for a certain period of time (respectively 30min, 1h, 3h, 6h, 12h, 24h, 48h).
[0120] 3. Take a 4×8 nitrocellulose membrane, drop 2 μL of samples onto the membrane sequentially, and place it at room temperature for 15 minutes.
[0121] 4. Place the film in an oven at 80°C and bake for 30 minutes.
[0122] 5. After taking it out from 80°C, block it with 5% milk, react at room temperature for 1 hour, and then wash it with PBST solution 3 times, 10 minutes each time.
[0123] 6. Incubate the NC membrane with the 5hmC antibody diluted in PBS solution, react overnight at 4°C, and then wash with PBST solution 3 times, 10 minutes each ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com