Label joint for detection of ultra-low-frequency gene mutation and application of label joint
A technology of tagging and sequencing, applied in the field of sequencing, can solve the problems such as the need to improve the tag connector and the low sensitivity of sample mutation detection, and achieve the effects of easy implementation, simple operation and accurate detection results.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
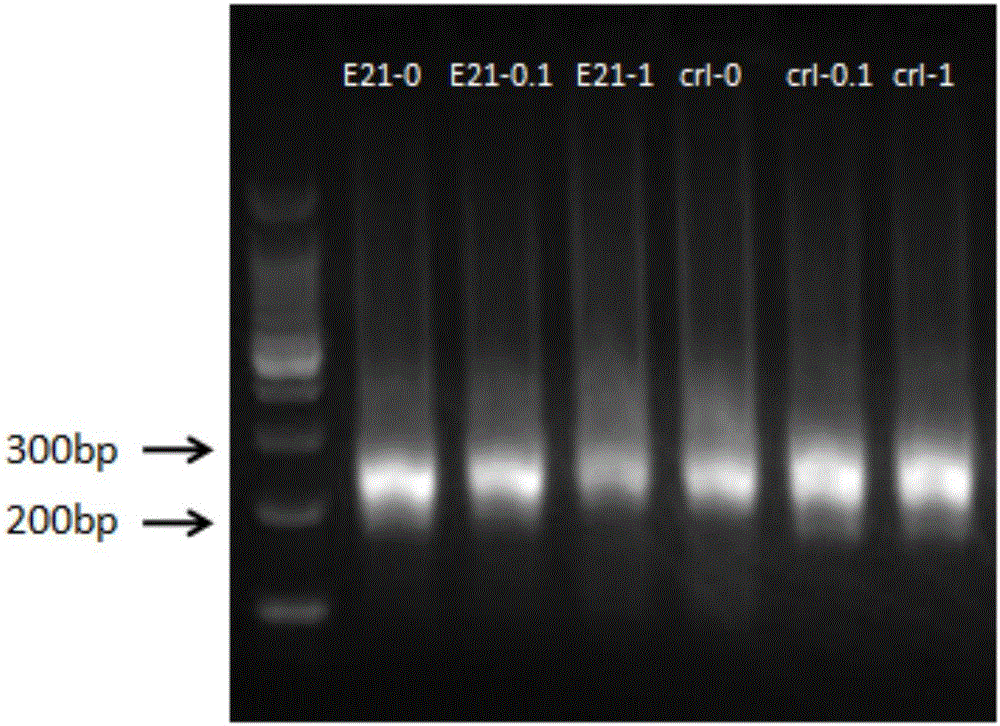
[0059] The library construction and the L858R mutation detection of exon 21 of the EGFR gene were performed on the sample using the tag adapter of the present invention (with the illumina Truseq adapter as the control), as follows:
[0060] 1. Label joint design of the present invention
[0061] Prelib-ADT1-S: CAAGCAGAAGACGGCATACGAGATTNNNNNNNNggaattaGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT (SEQ ID NO: 1),
[0062] Prelib-ADT2-S: CAAGCAGAAGACGGCATACGAGATNNNNNNNNatccggcGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT (SEQ ID NO: 2),
[0063] Prelib-ADT3-S: CAAGCAGAAGACGGCATACGAGATNNNNNNNNNcaggccgGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT (SEQ ID NO: 3),
[0064] Prelib-ADT-AS: pGATCGGAAGAGC,
[0065] The 5' end of the Prelib-ADT-AS sequence is modified with a phosphate group.
[0066] Prelib-ADT1-S, Prelib-ADT2-S, Prelib-ADT3-S (both are the first strands of the joint) are annealed with the Prelib-ADT-AS sequence to form double strands respectively, forming three joints of the present invention Prelib-A...
Embodiment 2
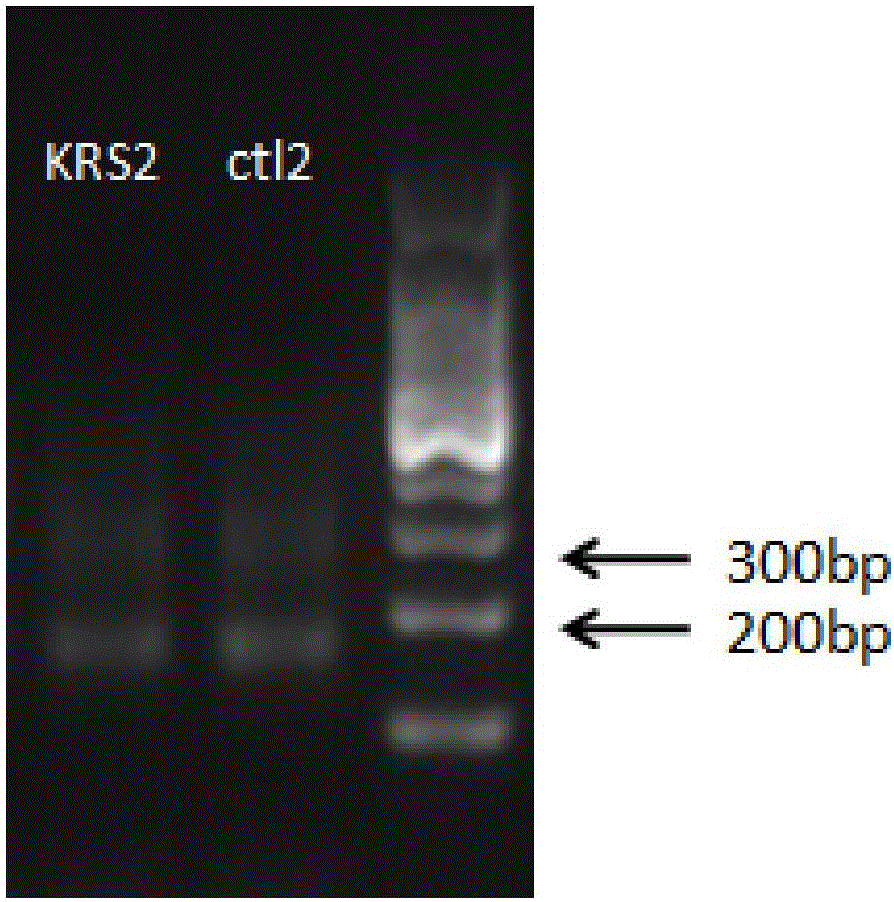
[0178] Using the tag adapter of the present invention (using the existing single-molecule tag-containing adapter as a control) to perform library construction and KRAS gene exon 2 G12D point mutation detection, the details are as follows:
[0179] 1. Label joint design of the present invention
[0180] Prelib-ADT1-S: CAAGCAGAAGACGGCATACGAGATTNNNNNNNNggaattaGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT (SEQ ID NO: 1),
[0181] Prelib-ADT-AS: pGATCGGAAGAGC, which is the modification of the phosphate group at the 5' end of the Prelib-ADT-AS sequence.
[0182] Prelib-ADT1-S and Prelib-ADT-AS sequences are annealed into double strands to form the linker Prelib-ADT1 of the present invention.
[0183] 2. The control group contains random single-molecule tag adapter design
[0184] According to the method described in the following literature, the design and manufacture of adapters containing single molecule tags: Scott R Kennedy, Michael W Schmitt.Detecting ultralow-frequency mutations by Dup...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com