Nucleic acid test strip for detecting cucumber green mottle mosaic viruses and application of nucleic acid test strip
A green mottled flower and leaf virus technology, applied in the direction of microbial-based methods, microbial measurement/inspection, biochemical equipment and methods, etc., can solve the problems of unfavorable grass-roots unit promotion and use, low sensitivity, and long time-consuming, etc., to achieve high sensitivity High efficiency, high detection efficiency, simple and convenient operation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0075] Example 1. Design and synthesis of primers
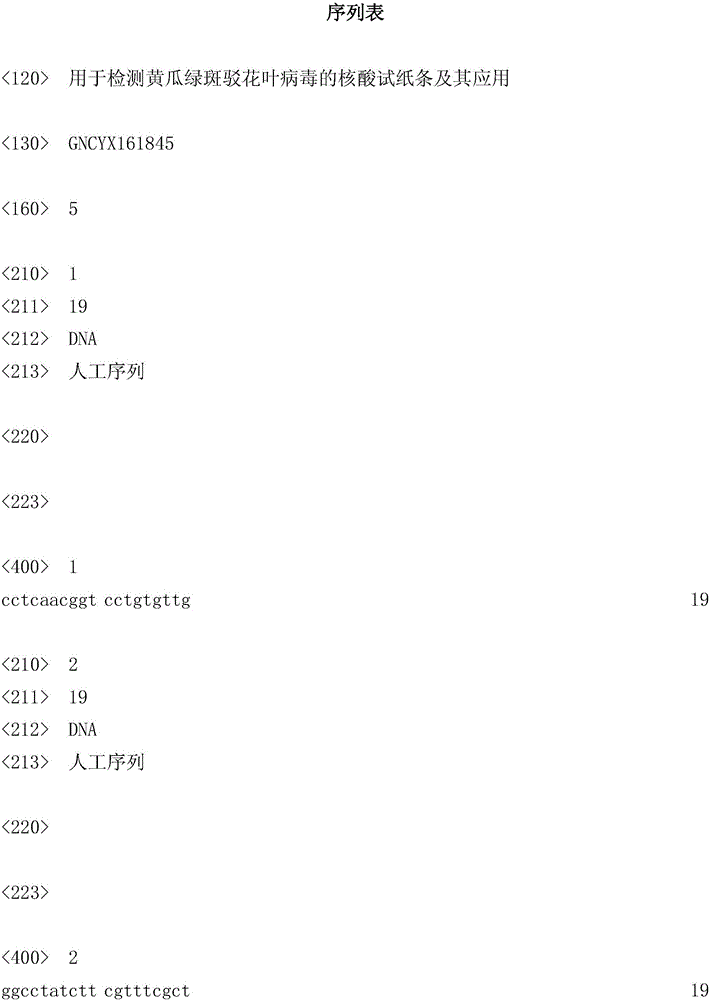
[0076] After a large number of sequence analysis, sequence design, manual screening optimization, and effect verification, a set of specific primers for the identification of cucumber green mottle mosaic virus was finally obtained. The specific primer set consists of primer CGMMV-BF, primer CGMMV-MBF, primer CGMMV-DF, primer CGMMV-CPR and primer CGMMV-BR.
[0077] CGMMV-BF (sequence 1): 5'-CCTCAACGGTCCTGTGTTG-3';
[0078] CGMMV-MBF (sequence 2): 5'-Biotin-GGCCTATCTTCGTTTCGCT-3';
[0079] CGMMV-DF (sequence 3): 5'-CTTAGCTCCACGGATACGC-3';
[0080] CGMMV-CPR (sequence 4): 5'-AACCTCAATGACCCTATTAGCGGCCTATCTTCGTTTCGCT-3';
[0081] CGMMV-BR (sequence 5): 5'-GTGGGATTGCTAGGATCTA-3'.
[0082] The 5' end of CGMMV-MBF was labeled with biotin, and the 5' end of CGMMV-DF was labeled with 6-FAM.
[0083] CGMMV-BF is a forward displacement primer. CGMMV-MBF is the detection probe 1. CGMMV-DF is the detection probe 2. CGMMV-CPR is a c...
Embodiment 2
[0084] Example 2, the establishment of the method
[0085] 1. Method for identifying whether the virus to be tested is cucumber green mottle mosaic virus
[0086] 1. Extract the total RNA of the virus to be tested.
[0087] 2. Using the total RNA extracted in step 1 as a template, the primer set designed in Example 1 was used to carry out reverse transcription cross primer isothermal amplification.
[0088] Reaction system (20 μl): primer CGMMV-CPR 1.5 μmol / L, biotin-labeled primer CGMMV-MBF 0.9 μmol / L at 5’ end, CGMMV-DF 0.9 μmol / L with 6-FAM label at 5’ end, Primer CGMMV-BF 0.3μmol / L, primer CGMMV-BR 0.3μmol / L, dNTP 0.4mmol, 10×Thermopol buffer 2μl, AMV reverse transcriptase 10 units, Bst DNA polymerase 8 units, MgSO 4 2 mmol, template 1 μl, and the balance is sterile water.
[0089] Reaction conditions: 59°C, 90min.
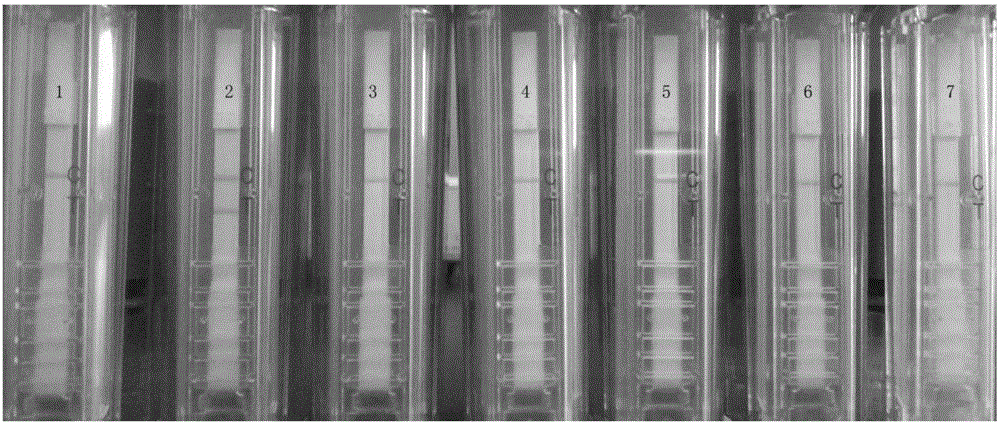
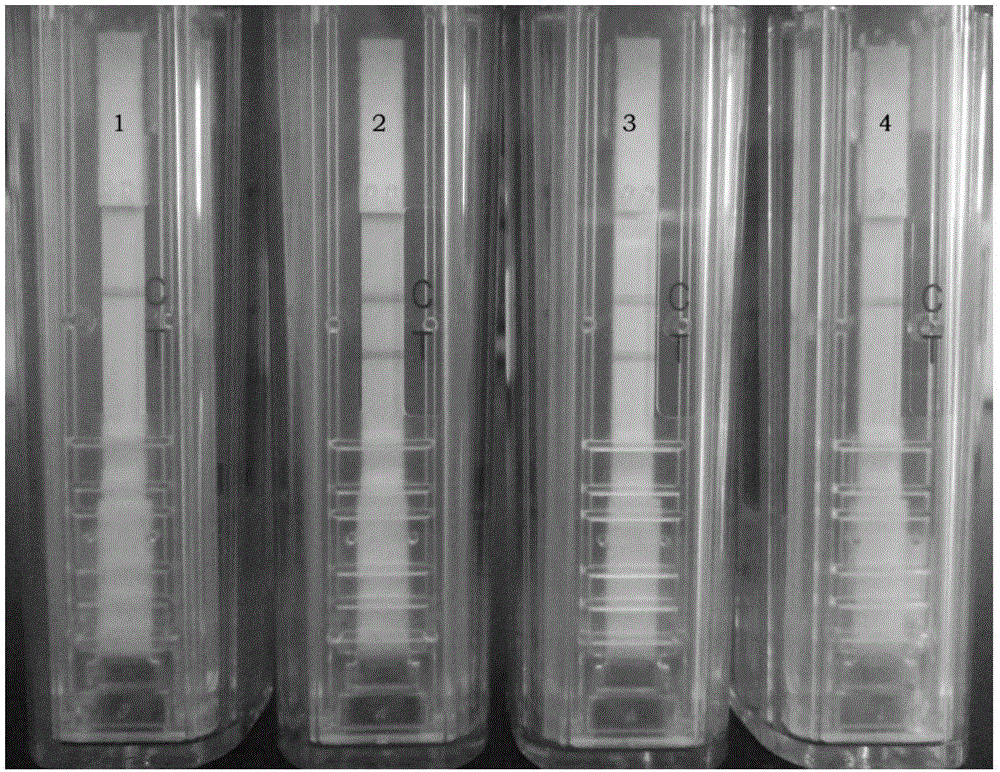
[0090] 3. Take the product of step 2 and use a disposable nucleic acid detection device for detection (operate according to the instructions). If both t...
Embodiment 3
[0095] Example 3. Specificity experiment
[0096] The viruses to be tested are as follows:
[0097] Cucumber green mottle mosaic virus (CGMMV): Reference: Chen Xiaoyu, Zhang Yongjiang, Li Guifen, etc. Research on the treatment effect of cucumber green mottle mosaic virus [J]. Plant Quarantine, 2010, 24(4 ): 16-19..
[0098] Cucumber mosaic virus (CMV): References: Li Guifen, Zhu Shuifang, Zhou Qun, etc. Identification of Cucumber mosaic virus isolates infecting Echinacea purpurea[J]. Plant Protection, 2007, 33(1) :54-56..
[0099] Pepper mild mottle virus (PMMoV-S): Reference: Zhang Yongjiang, Li Guifen, Ma Jie, etc. Detection of pepper mild mottle virus in pepper seeds[J].Anhui Agricultural Sciences,2007,35(14) :4228-4229..
[0100]Tobacco mosaic virus (TMV): References: Zhang Yongjiang, Ma Rongqun, Xin Yanyan, etc. Establishment of a barcode identification method for Tobacco mosaic virus [J]. Hubei Agricultural Science, 2015, 54(11): 2624-2627..
[0101] Tomato mosaic ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com