Primer pair for amplifying watermelon latent virus and application thereof
A latent virus and primer pair technology, applied in the field of RT-PCR reagents, RT-PCR kits, and biological detection, can solve the problems of lack of virus specificity and sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0059] Example 1: Design and synthesis of primers
[0060] Based on the genome of watermelon latent virus with GenBank accession number KY081285, after a large number of sequence analysis, sequence design, manual screening optimization, and effect verification, a specific primer pair for identifying watermelon latent virus was obtained for the conserved region RdRP encoded by dsRNA1. The specific primer pair consisted of CILCV-588-F and CILCV-588-R. The primer sequences are as follows:
[0061] CiLCV-588-F: 5'-TCCAGACGTTGGCTACACAC-3' (SEQ ID NO.1);
[0062] CiLCV-588-R: 5'-ATTGCGAACCTCTCAGGTGG-3' (SEQ ID NO.2);
[0063] Among them, CiLCV-588-F is the forward primer, and CiLCV-588-R is the reverse primer.
[0064] The theoretical amplification product size of the specific primer pair is 588bp, its nucleotide sequence is shown in SEQ ID NO.3 in the sequence listing, and the corresponding RNA sequence is shown in SEQ ID NO.4 in the sequence listing.
[0065] 5'-TCCAGACGTTGGCT...
Embodiment 2
[0067] Embodiment 2: establishment of identification method
[0068] One, the method for identifying whether the virus to be tested is watermelon latent virus
[0069] 1. Use the TRIZOL method or other kits that can be used to extract viral RNA to extract the total RNA of the sample to be tested according to the instructions.
[0070] 2. Using the total RNA extracted in step 1 as a template, use the primer pair designed in Example 1 to amplify by one-step RT-PCR.
[0071] The reaction system was as follows (25 μl): Primer CiLCV-588-F 0.5 μl (10 μmol / L), Primer CiLCV-588-R 0.5 μl (10 μmol / L), PrimeScript 1Step Enzyme Mix 1 μl, 2x 1stepBuffer (Dye Plus) 12.5 μl, Template 1 μL (that is, the total RNA extracted in step 1, the concentration is 100ng-200ng / μL), and the balance is made up to 25μl with RNase-free water. Among them, PrimeScript 1StepEnzyme Mix and 2x 1step Buffer (Dye Plus) are from Takara's PrimeScript TM One Step RT-PCR Kit Ver.2 (Dye Plus), Cat. No. RR057A.
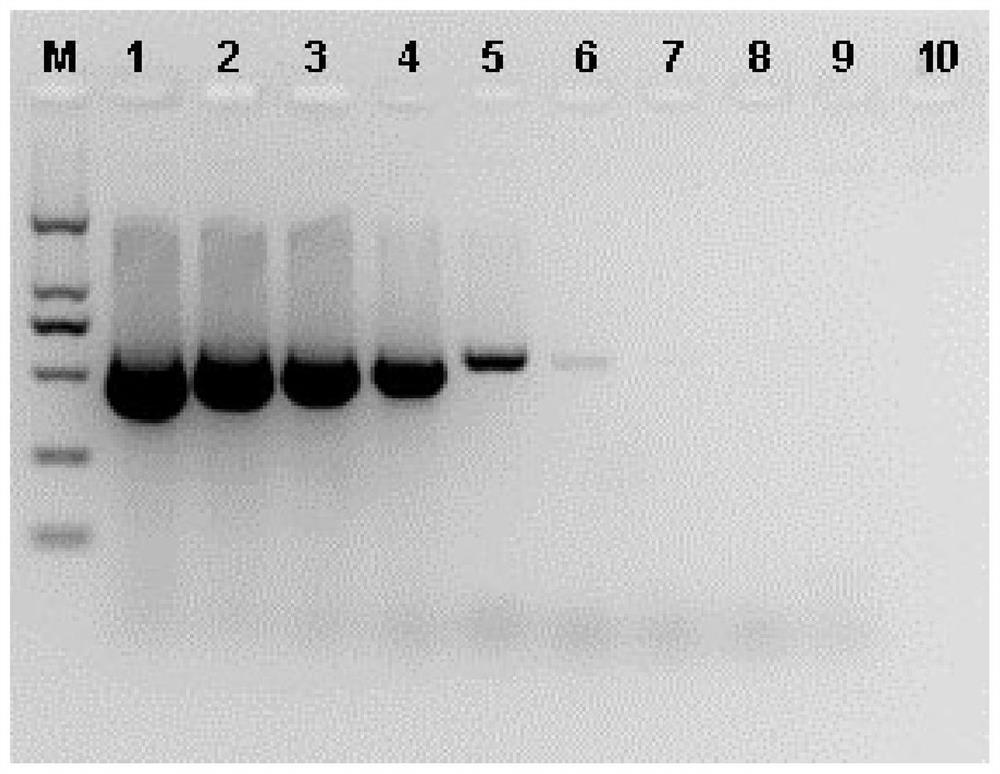
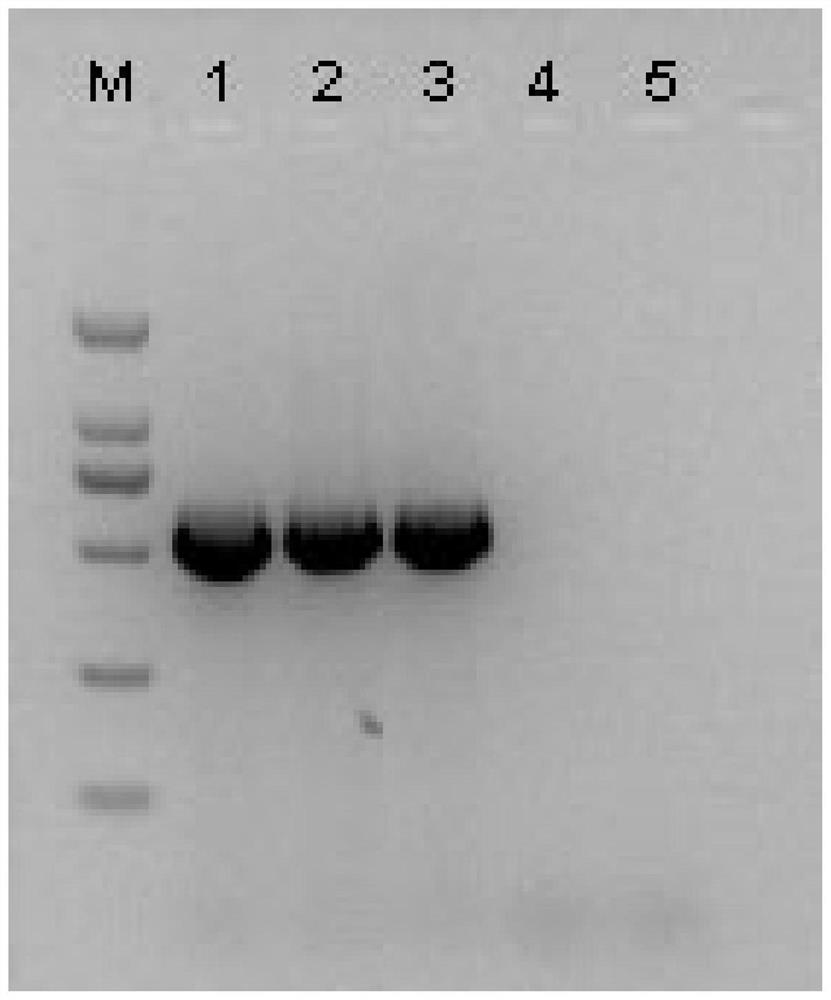
[0...
Embodiment 3
[0091] Embodiment 3: specificity experiment
[0092] The viruses to be tested are as follows:
[0093] Cucumber green mottle mosaic virus, cucumber mosaic virus, melon endogenous RNA virus, melon chlorotic virus, squash mosaic virus, tomato spotted wilt virus, watermelon mosaic virus, zucchini yellow mosaic virus.
[0094] The test samples of plants that have been confirmed to be infected with the above-mentioned viruses are tested according to "2. Method for identifying whether the plant material to be tested is infected with watermelon latent virus" in Example 2.
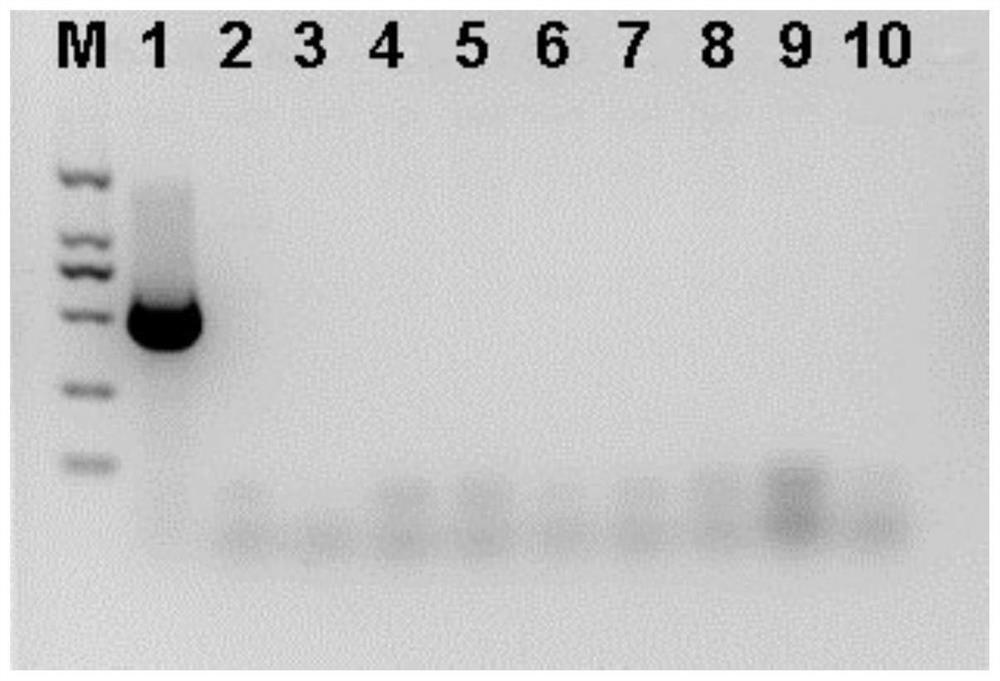
[0095] see results figure 1 . figure 1 Among them, lane 1 corresponds to the positive control watermelon sample containing watermelon latent virus; lane 2 corresponds to the detection of the amplification product of the watermelon sample containing cucumber green mottle mosaic virus; lane 3 corresponds to the detection of the amplification product of the watermelon sample containing cucumber mosaic virus. Ampli...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com