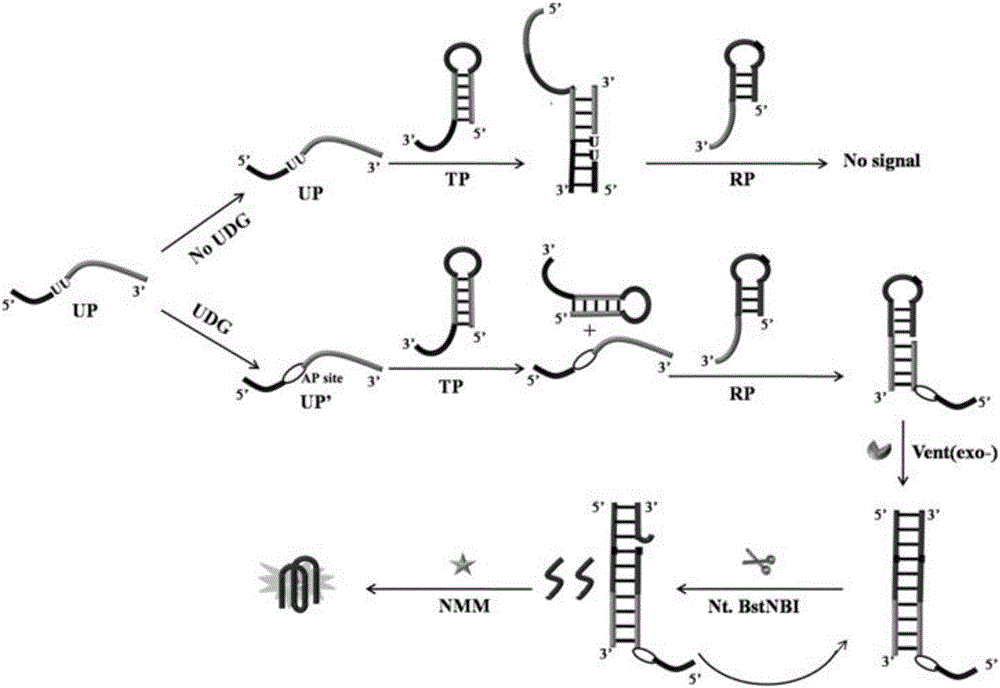
Method for detecting UDG (Uracil Dna Glycosylase) activity based on sticky end-mediated strand displacement reaction combined with polymerization incising isothermal amplification technology
A sticky end and reaction technology, which is applied in recombinant DNA technology, biochemical equipment and methods, microbial measurement/testing, etc., can solve the problems of limited sensitivity, weakened response, difficulty in distinguishing a small amount of mismatches, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
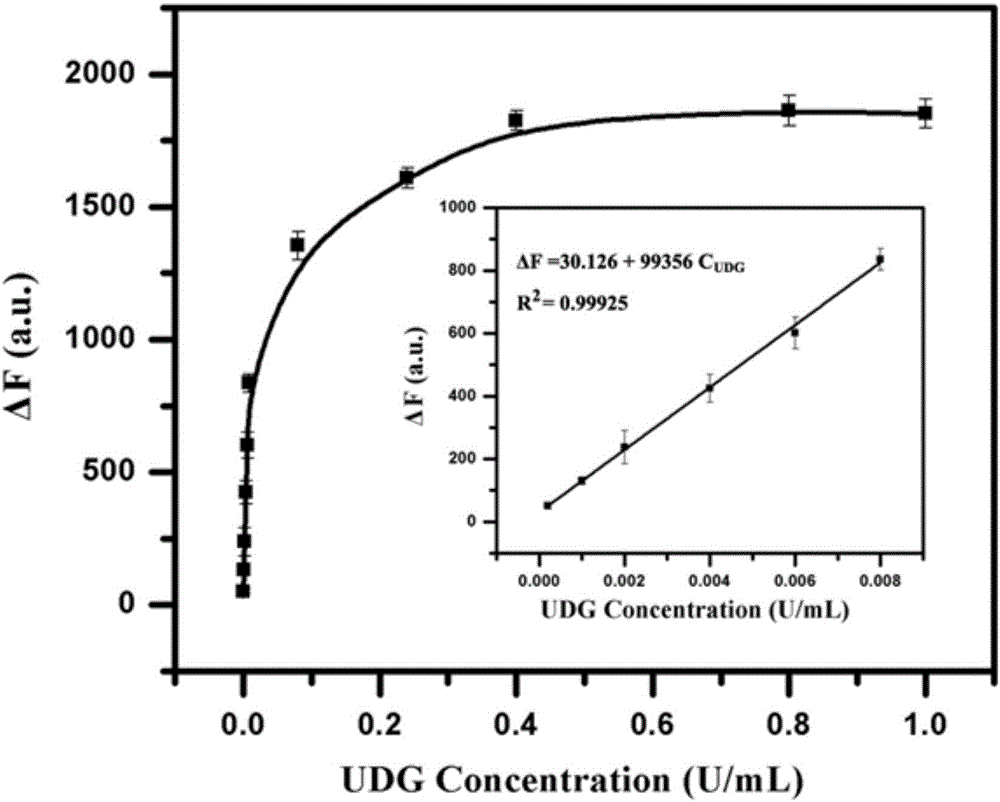
[0039] Embodiment 1 detects the activity of UDG
[0040] In order to obtain the corresponding hairpin structure, the hairpin probe TP containing sticky ends and the signal reporter probe RP were heated and denatured at 90 °C for 5 min, and the products were placed at 4 °C after the temperature was slowly lowered to room temperature. Save it for later use. Add 100nM-500nM recognition probe UP and different concentrations of UDG to 1×ThermoPol buffer solution (20mM Tris-HCl, pH 8.8, 10mM KCl, 10mM (NH 4 ) 2 SO 4 , 2mM MgSO 4 , 0.1% TritonX-100) to obtain a mixed solution with a final volume of 20 μL, and the solution was mixed and incubated at 37° C. for 20 min-60 min to complete the recognition reaction of UDG. Next, 150nM-1.80μM hairpin probe TP and 1.0μL 10×ThermoPol buffer solution (200mM Tris-HCl, pH 8.8, 100mM KCl, 100mM (NH4) 2 SO 4 , 20 mM MgSO 4 , 1% TritonX-100), to obtain a mixed solution with a final volume of 30 μL, and place the solution at 37° C. for 30 min...
Embodiment 2
[0041] Embodiment 2 The inventive method detects the UDG activity in the Hela cell lysate
[0042] Preparation of Hela cell lysate: Hela cell samples were pelleted by centrifugation (5min, 3000rpm, 4°C) and redispersed in lysis buffer (10mM Tris-Hcl, pH 7.0) by sonication on ice middle. The mixed solution was then centrifuged at 12,000 rpm for 30 min at 4°C to remove insoluble substances. The supernatant was collected and filtered through a 0.45 μm filter to generate a crude HeLa cell lysate.
[0043] 1.0 μL of HeLa cell lysate was used as the sample to be tested, wherein the concentration of the HeLa cell lysate was 1.0×10 per 1.0 μL of cell lysate 4 cells. Such as Figure 5 As shown, the buffer solution can only cause a very low fluorescence signal, while the HeLa cell lysate can cause a significantly enhanced fluorescence signal. In addition, when the UDG inhibitor UGI was added to the system containing HeLa cell lysate, the fluorescence intensity of the sensing system w...
Embodiment 3
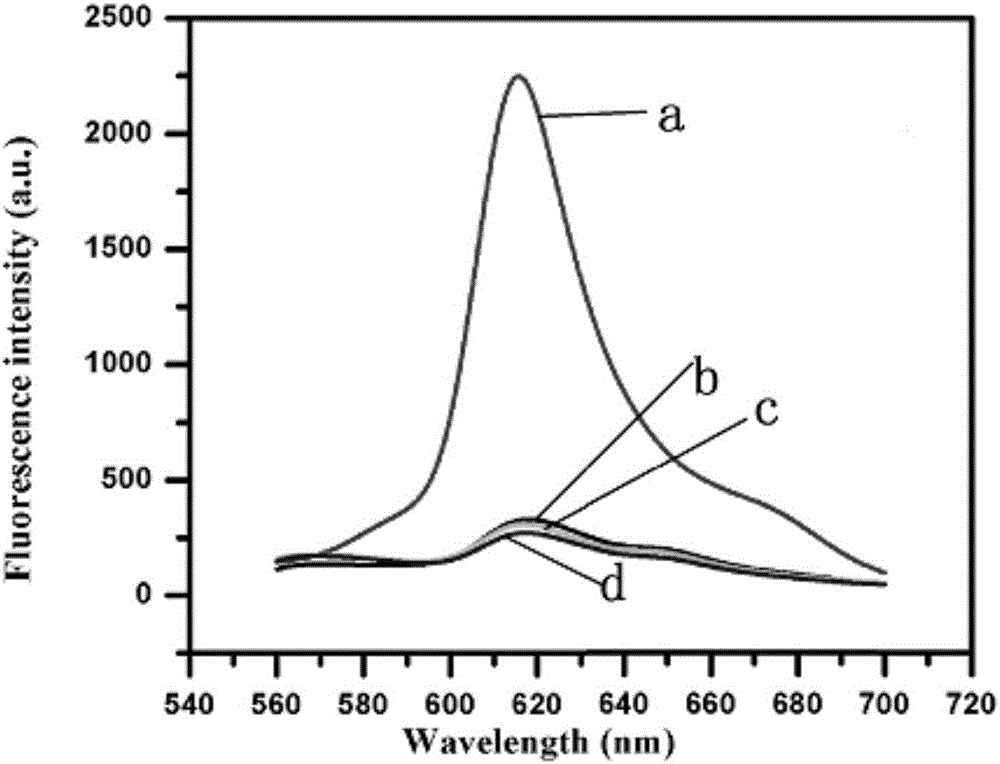
[0044] Example 3 Feasibility verification of detecting UDG activity
[0045] The feasibility of this method for detecting UDG activity was verified by fluorescence emission spectroscopy. Such as figure 2 As shown in curve d in , when the signal reporter probe RP is not included in the sensing system, the sensing system exhibits a very low fluorescence signal. This indicates that the generation of fluorescent signal is inseparable from the presence of probe RP. Such as figure 2 As shown in curve c in , when the recognition probe UP is not included in the system, the fluorescence signal of the sensing system is also very low. This indicates that in this method, the recognition of UDG by the sensing system is inseparable from the participation of the UP probe. Such as figure 2 As shown in curve b in , the sensing system still exhibits a very low fluorescence signal when UDG is absent. This is because the probe UP can generate TSDR with the hairpin probe TP, so that the p...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com