Genes highly expressed in tongue squamous carcinoma para-carcinoma tissue and applications of genes
A technology of tongue squamous cell carcinoma and genes, which is applied in the application field of genes in the diagnosis of tongue squamous cell carcinoma and the preparation of anti-tongue squamous cell carcinoma preparations, to achieve the effects of improving work efficiency, good stability, and rapid and accurate quantification
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0036] The collection and preparation of embodiment 1 material
[0037] Tongue squamous cell carcinoma and paracancerous tissue samples were taken from patients with tongue squamous cell carcinoma undergoing radical resection, and normal mucosal tissues were taken from patients with trauma. The specimens were collected within 30 minutes of the surgical procedure, and then collected under the guidance of pathologists. The sample size of each case is about 1.0cm×l.0cm×l.0cm. The cancer tissue is the central area of the tumor when the material is collected; the corresponding paracancerous tissue is the tissue 1-2cm away from the edge of the cancer tissue; the normal mucosa is excluding submucosal Epithelial tissue, the specimens were derived from surgical specimens of oral trauma. All specimens were placed in 1.5ml EP tubes and stored in liquid nitrogen tanks. All specimens were collected from patients who had not undergone any form of anti-tumor treatment and had no history ...
Embodiment 2
[0039] Embodiment 2 database and biological analysis method
[0040] RNA-seq read mapping
[0041] First, the low-quality reads were removed to obtain clean reads, and then TopHat v1.3.11 was used to match the clean reads with the UCSC H. sapiens reference genome (hg19), the human reference genome sequence (hgl9, updated on March 20, 2009) from Downloaded from UCSC database. The Hgl9 genome includes 25 reference sequences (reference assemblies) and 68 variable sequences (alternative assemblies). The pre-built index of H. sapiens UCSC hg19 version is downloaded from the TopHat homepage and used as a reference genome. When using TopHat to match the genome, each read (default to 20) is allowed to have multiple matching sites and a maximum of 2 mismatches . TopHat builds a library of possible splicing sites based on exon regions and GT-AG splicing signals, and maps reads that have not mapped to the genome to the genome based on these splicing site libraries. The system default...
Embodiment 3
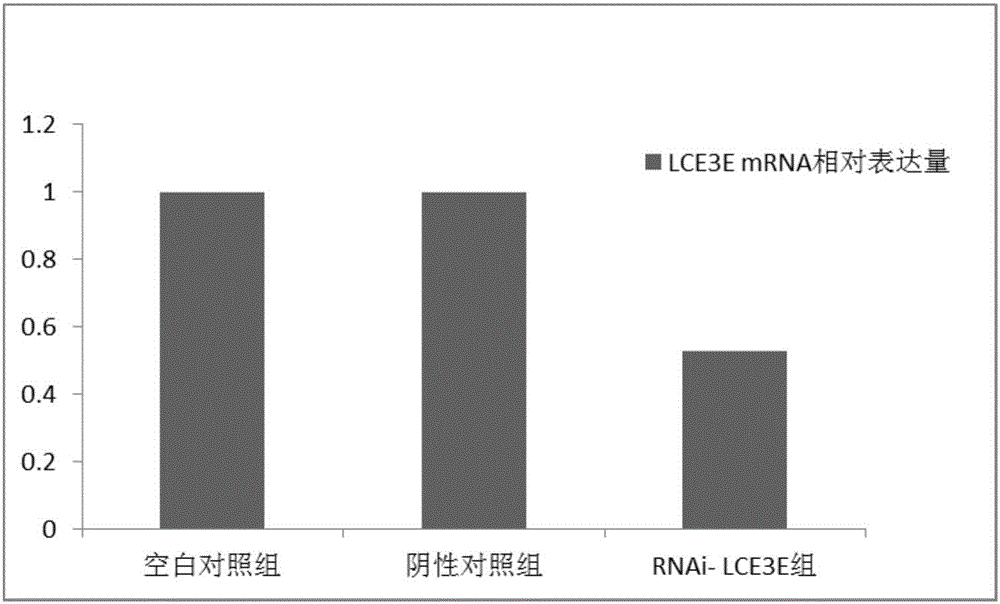
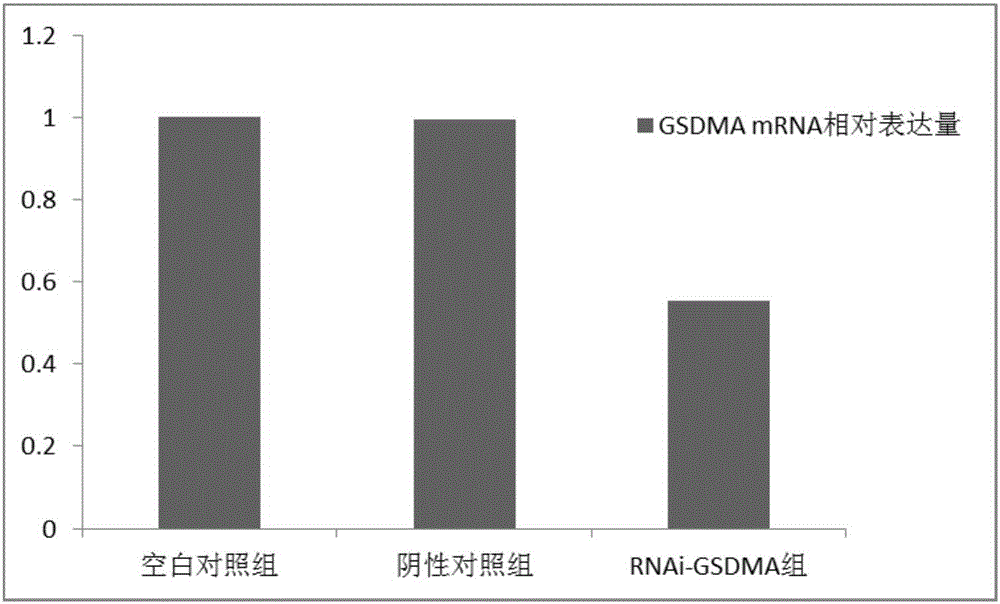
[0049] Example 3 Expression of LCE3E gene and GSDMA gene in tongue squamous cell carcinoma tissue and adjacent tissue
[0050] 1. Materials and methods
[0051] 1. Materials
[0052] See Example 1 for the collection criteria. 29 cases of tongue squamous cell carcinoma tissues and paracancerous tissues were collected and grouped and numbered.
[0053] 2. Method
[0054] 2.1 Extraction of RNA
[0055] The RNA was extracted according to Invitrogen's Trizol operating instructions, and stored in a -80°C ultra-low temperature freezer after extraction.
[0056] 2.2 Reverse transcription
[0057] By vazyme (HiScript TM Q RT Super Mix for qPCR) reverse transcription kit instructions. System: 4 μl of 4×gDNA wiper Mix; 2 μl of template RNA; RNase free ddH2O to make up to 20 μl. Gently pipette to mix, 42°C for 2 minutes.
[0058] Reverse transcription reaction system: 16 μl of the reaction solution in step 1 and 4 μl of 5×qRT SuperMix II, gently blow and mix with a pipette, incub...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com