Kit and method for detecting drug-resistant mutation site of helicobacter pylori
A drug-resistant mutation site, Helicobacter pylori technology, applied in biochemical equipment and methods, microbial measurement/inspection, etc., can solve the problems of time-consuming, complicated operation, and difficult cultivation of Helicobacter pylori
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
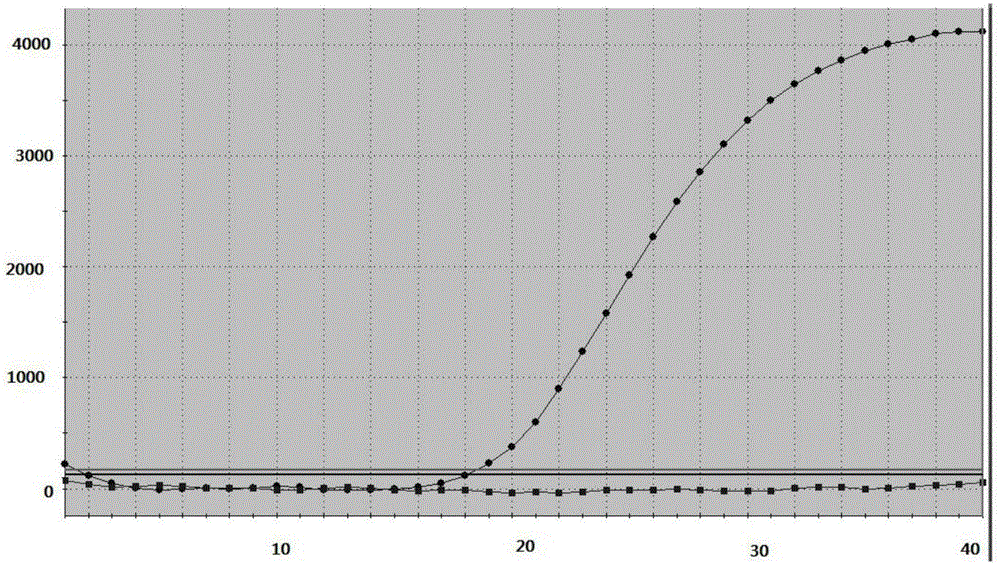
[0084] Example 1 Detection of the drug resistance of the kit of the present invention to the 259 site mutation of Helicobacter pylori gyrA gene
[0085]1. Determination of test samples
[0086] The clinical samples that were detected as Helicobacter pylori quinolone resistance by drug susceptibility test and proved to be mutated from A to T at the gyrA259 site by sequencing comparison were used as samples to be tested. Helicobacter pylori genomic DNA was extracted by column method or magnetic bead method, and stored at -80°C after aliquoting.
[0087] 2. Design of primers and probes:
[0088] Multiple pairs of primers and probes are designed, and the length of the primers is generally 15-30 bases.
[0089] The A259T primer probe and blocking sequence of the gyrA drug-resistant mutation site of Helicobacter pylori quinolone gene are as follows:
[0090] Upstream primer 259-F: 5'ACCACCCCCCATGGCGATT3'
[0091] Downstream primer 259-R: 5'CGCCATCAATAGAGCCAAAGTG3'
[0092] Prob...
Embodiment 2
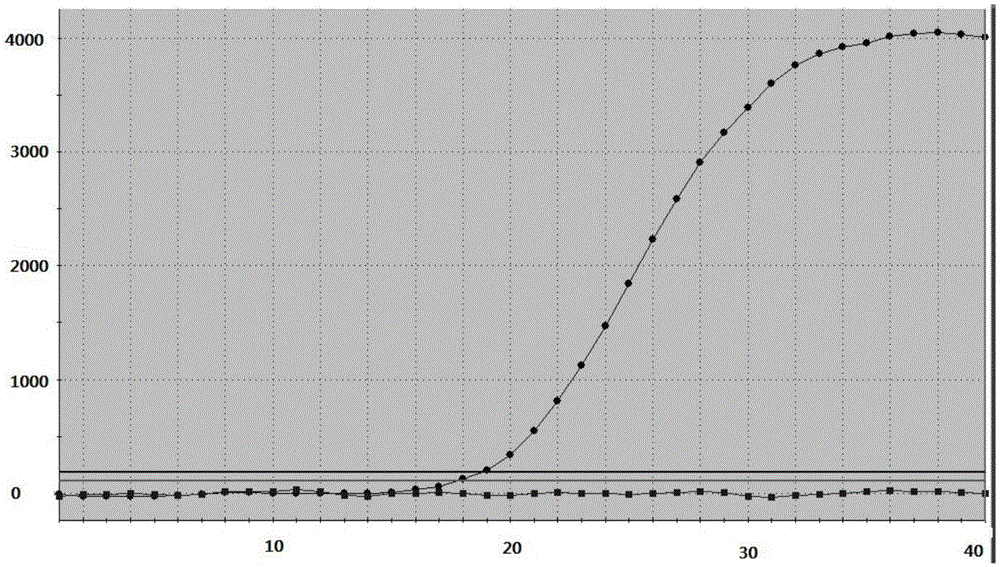
[0109] Example 2 Detection of the drug resistance of the kit of the present invention to the 261 site mutation of Helicobacter pylori gyrA gene
[0110] 1. Determination of test samples
[0111] The clinical samples that were detected as Helicobacter pylori quinolone resistance by drug susceptibility test and proved to be mutated from T to G at the gyrA261 site by sequencing comparison were used as samples to be tested. Bacterial genomic DNA was extracted by column method or magnetic bead method, and stored at -80°C after aliquoting.
[0112] 2. Design of primers and probes:
[0113] Multiple pairs of primers and probes are designed, and the length of the primers is generally 15-30 bases.
[0114] In this example, the T261G primer probe and blocking sequence of the Helicobacter pylori quinolone gene gyrA drug-resistant mutation site are as follows:
[0115] 261-F: 5'ACCACCCCCCATGGCGATTAG3'
[0116] 261-R: 5'CGCCATCAATAGAGCCAAAGTG3'
[0117] 261-P: 5'HEXGAGAATGGCGCAAGATTTT...
Embodiment 3
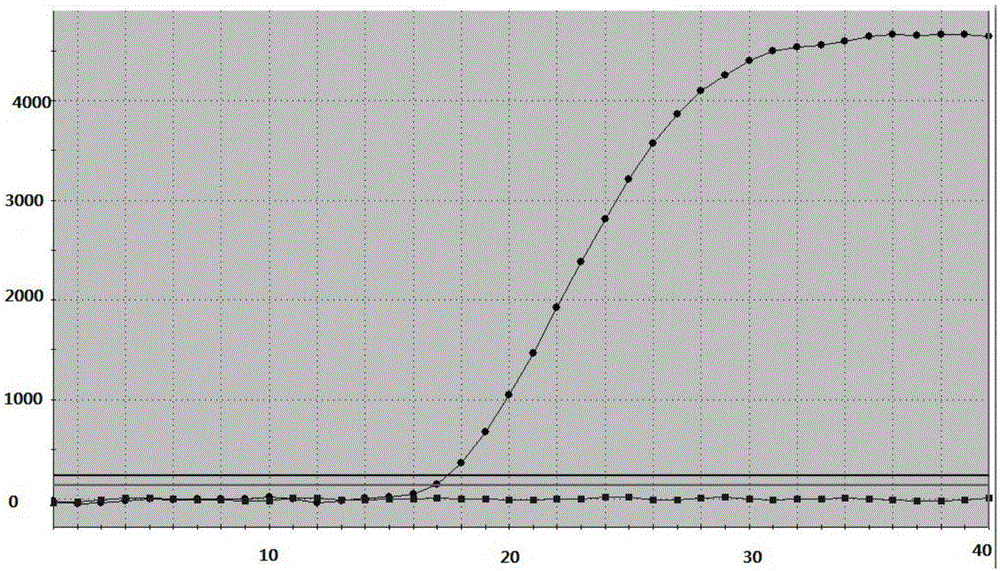
[0135] Example 3 Detection of the drug resistance of the kit of the present invention to the 271 site mutation of Helicobacter pylori gyrA gene
[0136] 1. Determination of test samples
[0137] The clinical samples that were detected as Helicobacter pylori quinolone resistance by drug susceptibility test and proved to be mutated from G to A at the gyrA271 site by sequencing comparison were used as samples to be tested. Bacterial genomic DNA was extracted by column method or magnetic bead method, and stored at -80°C after aliquoting.
[0138] 2. Design of primers and probes:
[0139] Multiple pairs of primers and probes are designed, and the length of the primers is generally 15-30 bases.
[0140] In this example, the G271A primer probe and blocking sequence of the gyrA drug-resistant mutation site of Helicobacter pylori quinolone gene are as follows:
[0141] 271A-F: 5'CCATGGCGATAATGCGGTTTTTA3'
[0142] 271A-R: 5'CGCCATCAATAGAGCCAAAGTG3'
[0143] 271A-P: 5'HEXGAGAATGGCGC...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Sensitivity | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com