GeXP analysis method for synchronous detection of vibrio cholera and vibrio parahaemolyticus in food
A technology for Vibrio hemolyticus and Vibrio cholerae, which is applied in the directions of microorganism-based methods, biochemical equipment and methods, and the determination/inspection of microorganisms, which can solve the problems of simultaneous detection of Vibrio cholerae, Vibrio parahaemolyticus, etc. , to avoid competitive effects, high sensitivity, and improve sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0026] Embodiment 1, primer design
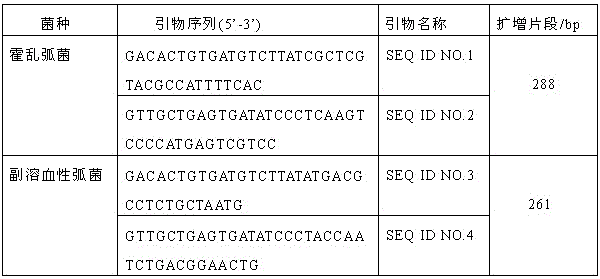
[0027] The ompW gene of Vibrio cholerae and the toxR gene of Vibrio parahaemolyticus were selected as specific genes for amplification, and specific primers were designed using eXpressDesigner. The primer sequences and product sizes are shown in Table 1.
[0028] Table 1
[0029]
[0030] The universal primers are a pair of non-biological oligonucleotide sequences, and the 5' ends of the upstream primers are labeled with cy5 fluorescence. The sequences are shown in Table 2.
[0031] Table 2
[0032] Primer name Primer sequence (5'-3') SEQ ID NO.5 GACACTGTGATGTCTTAT SEQ ID NO.6 GTTGCTGAGTGATATCCCT
[0033] The above primers were synthesized by Sangon Bioengineering (Shanghai) Co., Ltd.
Embodiment 2
[0034] Example 2, using the GeXP method to simultaneously detect Vibrio cholerae and Vibrio parahaemolyticus in aquatic products (yellow croaker).
[0035] Take 25g of samples to be tested from different parts of yellow croaker and inoculate them in 225mL of 3% sodium chloride alkaline peptone water, place them in a biochemical incubator at 37°C±1°C and incubate for 18-24 hours.
[0036] Nucleic acid extraction: Take 1.5 mL of the enrichment culture solution and place it in a centrifuge tube, centrifuge at 12,000 rpm for 3 minutes and discard the supernatant. A general-purpose commercial Gram-negative bacterial DNA extraction kit was selected, and the nucleic acid of the culture was extracted according to the steps specified in the kit instruction manual, and stored at -70°C for future use.
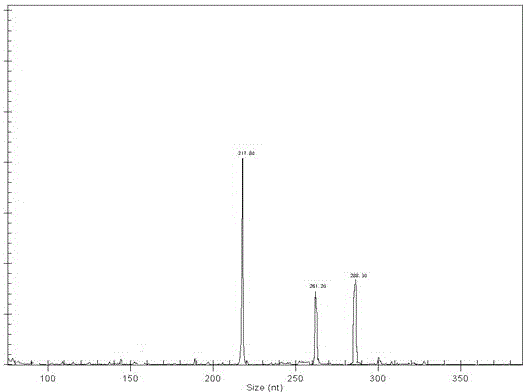
[0037]Configuration and amplification parameters of the PCR amplification system: 25 μL reaction system, containing 0.2 μL each of specific primers SEQ ID NO.1 and SEQ ID NO.2 (0.1 mM), S...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com