Kit for detecting DNA mismatch repair systems and application thereof
A technology of mismatch repair and kit, applied in the field of DNA mismatch repair system, can solve the problems of discrepancy in results, high price, inaccurate diagnosis, etc., to reduce human subjective judgment and standardization, high sensitivity and accuracy, and improve The effect of detection efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
experiment example 1
[0039] Experimental example 1 PCR amplification reaction
[0040] 1. PCR reaction system
[0041] Table 1: Reaction specific components
[0042]
[0043] The dNTPs in Table 1 above include dATP, dGTP, dTTP and dCTP, and the use concentration of dATP, dGTP, dTTP and dCTP is 1 mM.
[0044] The primer pair mixture used refers to a mixture containing the microsatellite site of the present invention and its upstream primer and downstream primer, and the final concentration of each primer pair (upstream primer and downstream primer) is 0.1-1 μM. As shown in Table 2, the specific components of the two mixtures.
[0045] Table 2: Specific components of the two mixtures
[0046]
[0047]
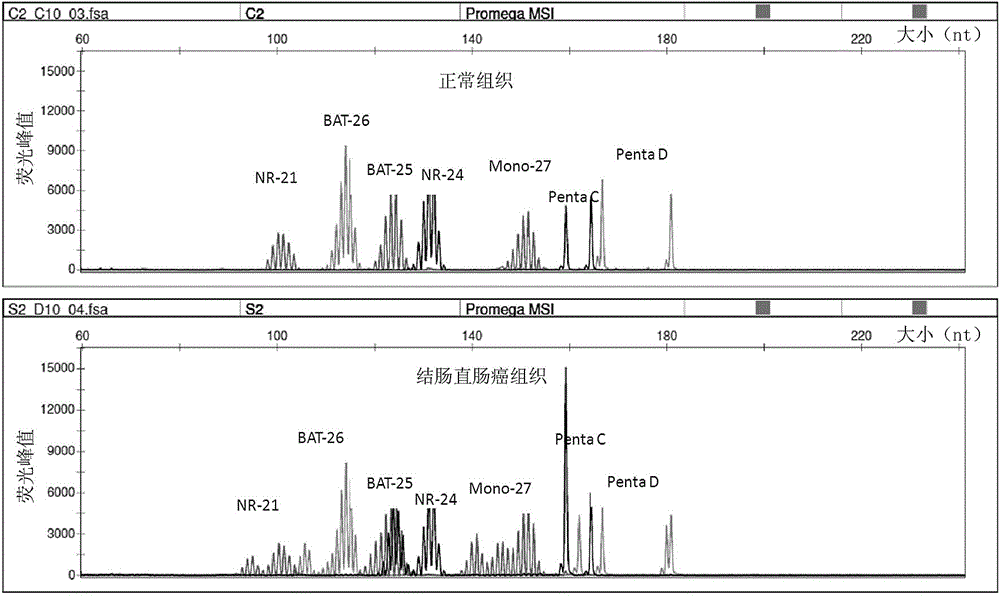
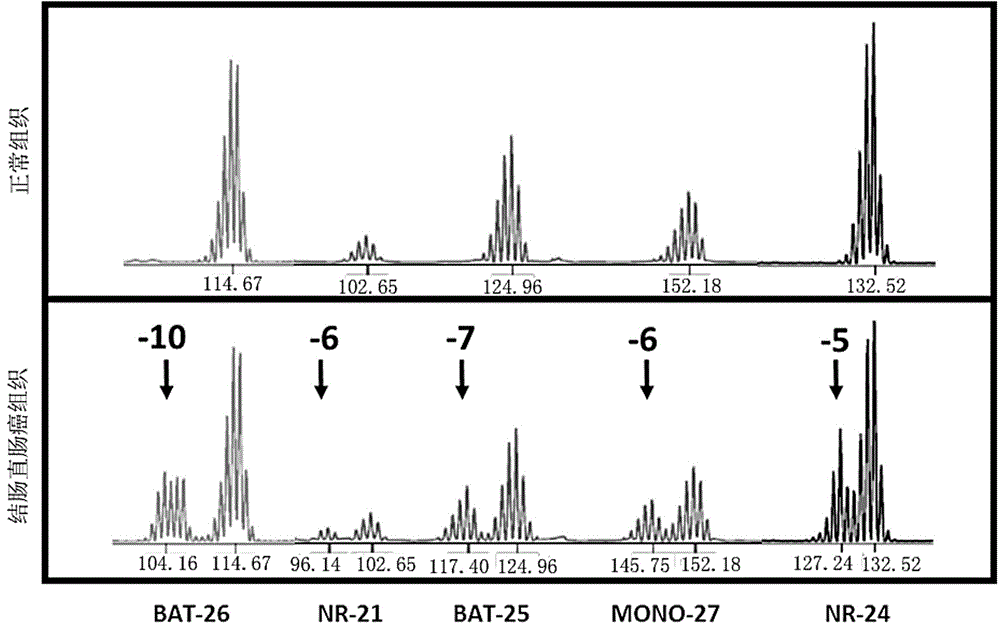
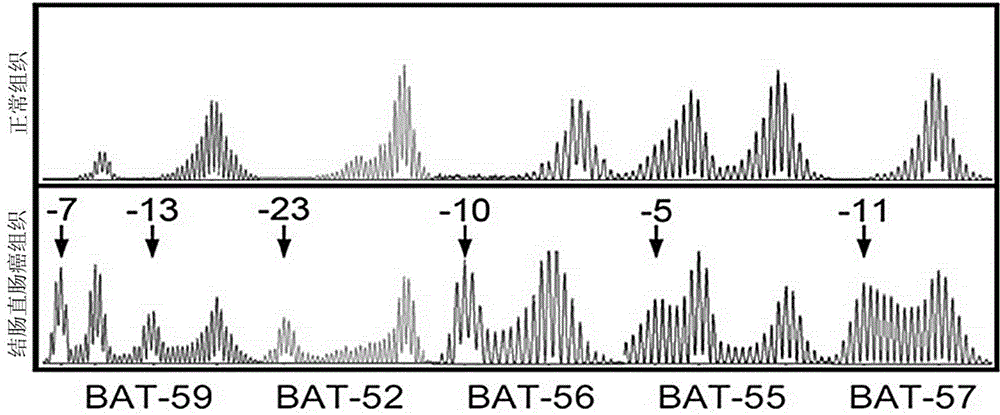
[0048] In the above table 2, the 5'end fluorescent label JOE is 6-carboxy-4',5'-dichloro-2',7'-dimethoxyfluorescein, FL is Fluorescein, ET-TMR is Energy Transfer-carboxy-tetramethylrhodamine, " / "Means no fluorescent labeling, and the 5'end is hydroxyl. Among them, the microsatellite sites Penta C and Penta ...
experiment example 2
[0054] Experimental example 2 Capillary electrophoresis experiment
[0055] Take the amplified PCR product, add internal standard (Promega company, internal molecular standard ILS500) and formamide, dry bath at 95 ℃ for 3 minutes, and ice water bath for 3 minutes. Load the sample on the ABI 3500Dx instrument, open the 3500Dx data collection software, edit the sample template, import the fragment analysis program, and click Run. Specific steps are as follows:
[0056] (1) Prepare the sample for loading: 1μL of the obtained PCR product, add 10μL of formamide, and 1μL of DNA molecular standard internal standard ILS500;
[0057] (2) Capillary electrophoresis: Set the capillary electrophoresis program according to the ABI3500Dx instrument manual, as follows:
[0058] A. Prepare the instrument; B. Preheat the heating furnace to 60°C; C. Prepare the sample reaction plate and load it into the instrument; D. Check the instrument status; E. Create or import a reaction plate; F. Assign the cont...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com