Method for knocking out drug resistance gene mcr-1 through CRISPR-Cas9 in vitro and specialized cell penetrating peptide thereof
A drug resistance gene, mcr-1 technology, applied in the field of microbial genetic engineering and biology, to achieve the effect of broad application prospects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0047] Example 1 Construction of the CRISPR-Cas9 system for the mcr-1 gene
[0048] 1. Compare the mcr-1 gene sequence to find a relatively conserved region. The mcr-1 gene-specific targeting sequence in this region is shown in SEQ ID NO. 1. The targeting sequence is designed with sgRNA and a sgRNA, its corresponding DNA sequence information is shown in SEQ ID NO.5.
[0049] 2. Construction of pCas9-mcr:
[0050] (1) Design and synthesize the sgRNA DNA sequence oligo DNA that recognizes the mcr-1 gene according to SEQ ID NO.5;
[0051] (2) Perform gradient cooling and annealing on the synthesized oligo DNA sequence, the specific steps are:
[0052] Mix the synthesized oligo DNA (100 μM, 1 μL) with 10xPCR Buffer (1 μL), then add 10 μL of water to make up the system; then denature at 95°C for 5 minutes, and then cool down to 25°C at a rate of 2°C per minute to complete the reaction; anneal After that, a synthetic oligo DNA hybrid duplex is formed.
[0053] (3) Use BsaI to di...
Embodiment 2
[0063] Example 2 Penetrating peptide CPP5a in Staphylococcus aureus JS17
[0064] The test steps are as follows:
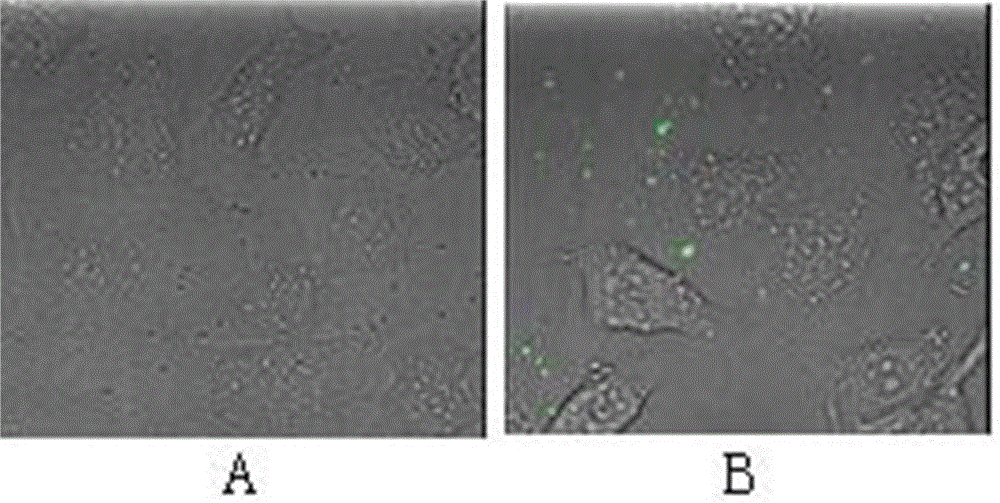
[0065] 1. Synthesize the cell penetrating peptide CPP5a according to SEQ ID NO.2, and use Alexa Fluor R 488 ProteinLabeling Kit for labeling (the operation steps were carried out in strict accordance with the instructions), and the protein after labeling was named CPP5a-488; the labeling compound can be combined with the N-terminal NH of the protein 2 - The residues are linked so that the labeled protein emits green fluorescence under laser excitation.
[0066] 2. Add Staphylococcus aureus JS17 to the TSB medium, and then add CPP5a-488 at a final concentration of 0.1 mg / ml, co-cultivate at room temperature for 1 hour, and wash off the recombinant protein with PBS.
[0067] 3. Take the MAC-T cells infected with Staphylococcus aureus JS17 co-cultured in step (2) for 1 hour as the experimental group (GFP-CPP5a), and set up MAC-T cells infected with Staphylococcus...
Embodiment 3
[0068] Example 3 Gel retardation experiment of penetrating peptide CPP5a and sgRNA-Cas9 expression vector pCas9-mcrDNA The binding of cell penetrating peptide CPP5a to DNA was analyzed by electrophoretic mobility. Migration is affected, ie there is a lag compared to the distance that DNA alone migrates.
[0069] Experimental group (12 μL): DNA of vector pCas9-mcr 250 ng, vector pCas9-mcr obtained in step (4) of Example 1 45 μg, penetrating peptide obtained in step (5) of Example 1 45 μg, ddH 2 0 to make up 12 μL;
[0070] Control group (12 μL): DNA of vector pCas9-mcr 250 ng, vector pCas9-mcr obtained in step (4) of Example 1 45 μg, ddH 2 0 to make up 12 μL;
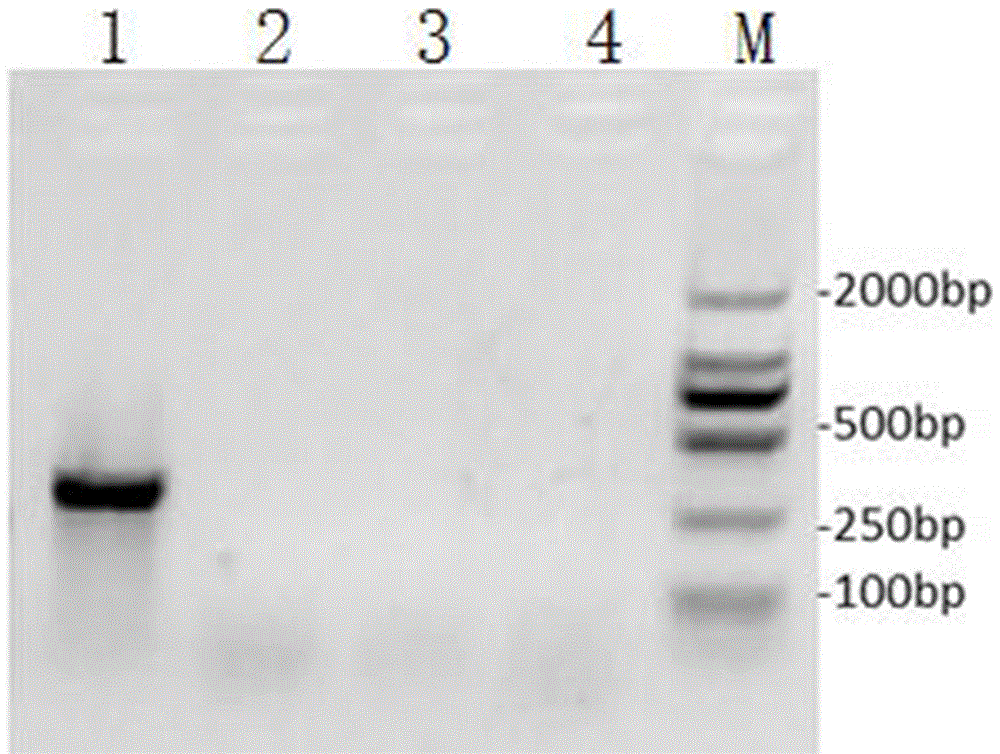
[0071] The control group and the experimental group were incubated at room temperature for 30 minutes, respectively. After incubation, run electrophoresis with 0.8% agarose gel at a voltage of 120V. After electrophoresis, place the electrophoresis gel in a gel imaging system, observe and record under ultraviolet light...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com