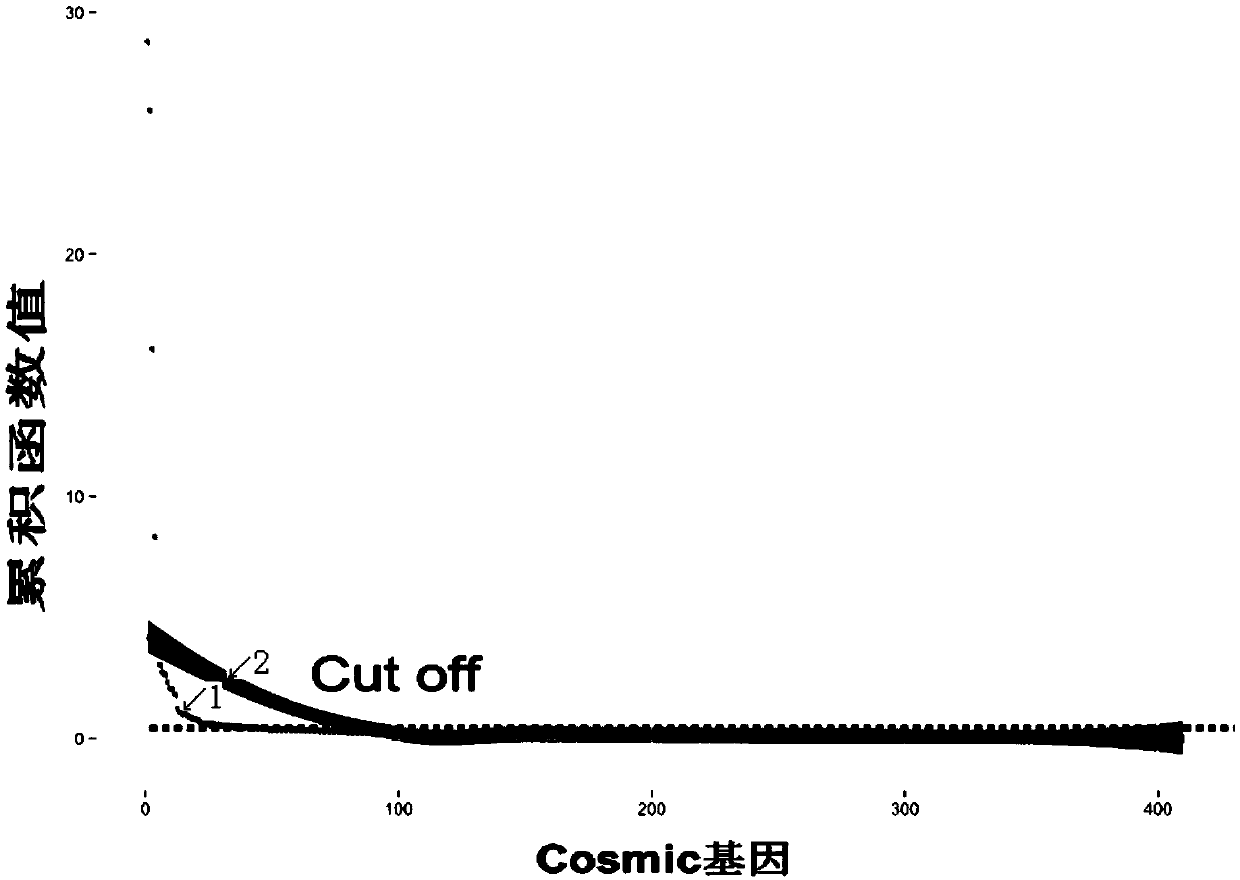
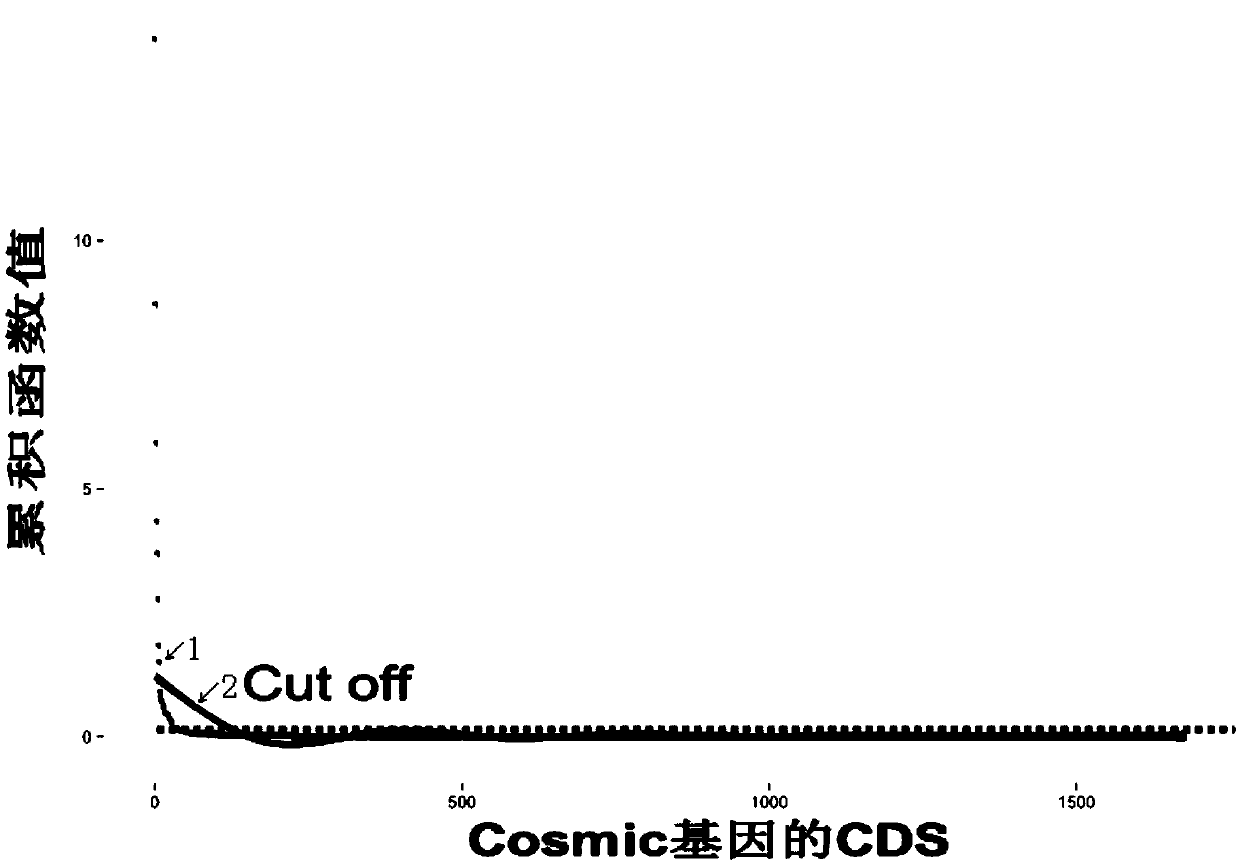
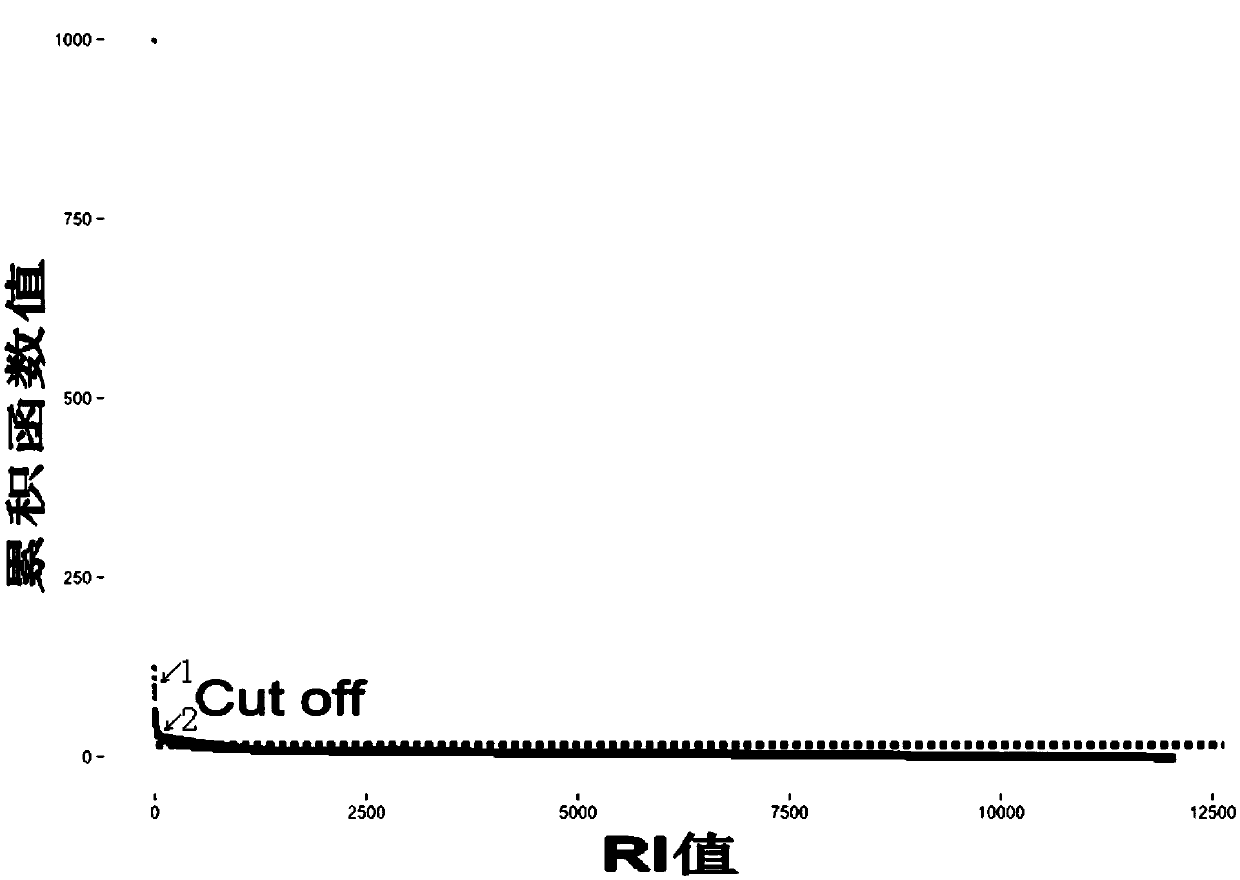
Specific tumor probe region design method and device and probe for target region capture high-throughput sequencing
A technology of target region capture and design method, applied in the field of tumor probe design, can solve the problems of lack of pertinence, high cost, and inability to conduct large-scale research on liver cancer patients in gene chip design, reducing sequencing time and cost, and shortening detection cycle. , the effect of reducing the sequencing range
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0066] According to the embodiment of the present invention, the whole exome data of 192 patients with liver cancer in the TCGA database were selected to evaluate a group of probes of the present invention, and the results showed that 107 patients (55%) had severe mutations in this region, and at the same time It was found that in the designed specific liver cancer chip region (LIVCAN_REG), the average 1Kb region contained at least one severe mutation, and a 211Kb region was randomly selected on the human genome for random data simulation, and the results showed that the randomly selected region averaged 1Kb Only about 0.0123 mutations related to liver cancer are contained, so it can be clearly seen that the chip region designed by the present invention is related to liver cancer, and the specificity of the chip is verified. At the same time, it shows that the probe (LIVCAN_SEQ) designed by the present invention has a very high detection rate of at least a part of the liver can...
Embodiment 2
[0068] According to the method for determining the nucleic acid sequence of the gene coding region of the sample to be tested according to the present invention, in this embodiment, a chip (chip508) data containing 508 gene coding regions is used for verification. chip508 covers currently known and various types of Cancer-related and reported mutated genes, as well as some genes related to chemotherapy and targeted drugs. Through the combination of target sequence capture technology, high-throughput sequencing and biological information analysis, 508 genes in the collected DNA samples of blood and cancer tissues were detected, and then the liver cancer-specific region (LIVCAN_REG) obtained in the present invention was analyzed. Analysis, to obtain the mutation information of the corresponding gene, and horizontally compare the obtained mutation region with the designed specific liver cancer region to verify the specificity of the design of the present invention.
[0069] Among...
Embodiment 3
[0210] According to the method for determining the nucleic acid sequence of the gene coding region of the sample to be tested according to the present invention, in this embodiment, a chip (chip508) containing 508 gene coding regions is also used for verification. Through sequence capture technology, high-throughput sequencing, and methods combined with bioinformatics analysis, 508 genes were detected in blood cell DNA samples and cancer tissue samples collected from 3 liver cancer patients, and then the liver cancer specificity obtained in the present invention The region is analyzed to obtain the mutation information of the corresponding gene, and the obtained mutation region is horizontally compared with the designed specific liver cancer region (LIVCAN_REG) to verify the specificity of the designed LIVCAN_REG region of the present invention.
[0211] Among them, the sample to be tested is the patient information as shown in Table 10:
[0212] Table 10
[0213] pa...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com