Recombinant escherichia coli for cell surface display PET lytic enzyme, construction and application
A technology for recombining Escherichia coli and cell surface, applied in the field of genetic engineering of enzymes, can solve the problems of slow degradation rate, long growth cycle, popularization and application of restriction enzymes, etc., and achieves limited solution ability, simple preparation method and use, and biocatalytic activity. high effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
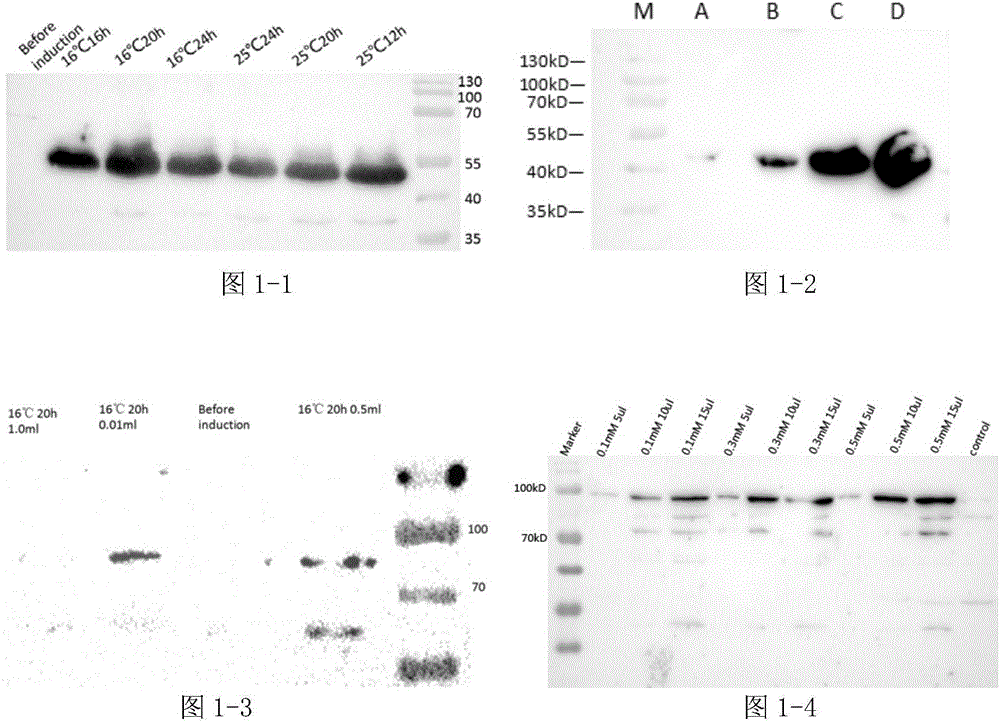
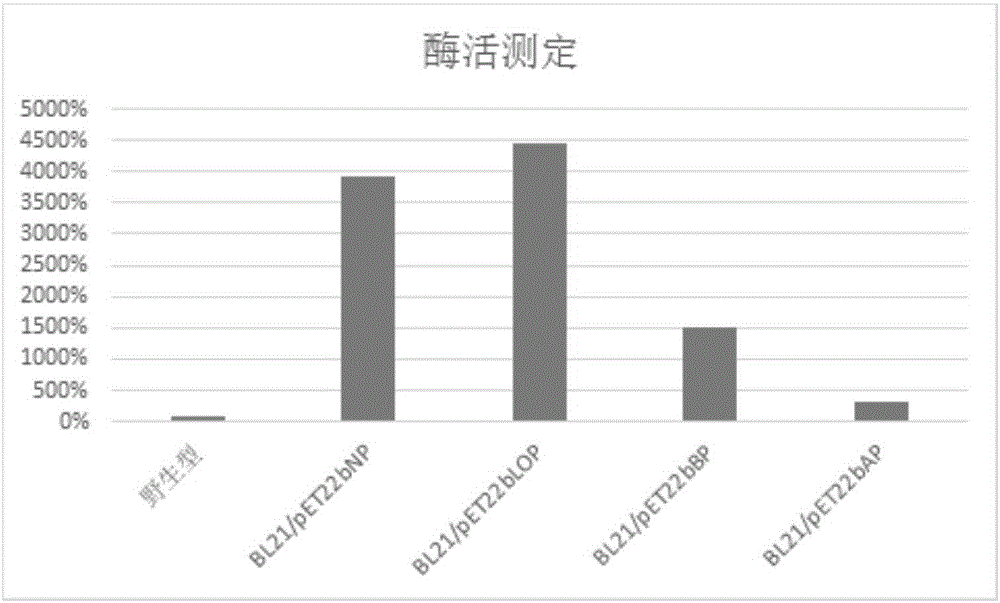
[0033] Example 1: Construction and application of recombinant Escherichia coli (BL21 / pET22bNP) displaying PET decomposing enzyme on the cell surface
[0034] Chemically synthesize the inaK-N gene as shown in SEQ ID No.2 as the anchor protein, and chemically synthesize the PETase gene as shown in SEQ ID No.1;
[0035] Primers were designed using the inaK-gene sequence and PETase gene sequence as templates.
[0036] Upstream primer inaK-NF1 5'G GAATTC CATATGACACTTGATAAGGCGC 3' (the underlined part is the NdeI restriction site)
[0037] Downstream primer inaK-NR1 5'ATAAGGATTGGTTTGAGTCTGTAAGTTCTGAGGGG 3'
[0038] Upstream primer inaK-NF2: 5'CAGAACTTACAGACTCAAACCAATCCTTATGCCC 3'
[0039] Downstream primer inaK-NR2:5'CC CTCGAG GCTACAGTTGGCGGTACGA 3' (the underlined part is the XhoI restriction site)
[0040] The target genes inaK-N and PETase were obtained by PCR amplification using artificially synthesized inaK-N gene and PETase gene as templates. The PCR reaction parameter...
Embodiment 2
[0061] Example 2: Construction and application of recombinant Escherichia coli (BL21 / pET22bLOP) displaying PET decomposing enzyme on the cell surface
[0062] Chemically synthesize the anchor protein Lpp-OmpA gene shown in SEQ ID No.3, and chemically synthesize the PETase gene shown in SEQ ID No.1;
[0063] Primers were designed using Lpp-OmpA gene sequence and PETase gene sequence as templates.
[0064] Upstream primer LOP-F1 5'GGTCTTC CATATG AAAGCTACTAAACTGGTAC 3' (the underlined part is the NdeI restriction site)
[0065] Downstream primer LOP-R1 5'CACTACCTCCACCACCGTTGTCCGGACGAGTG 3'
[0066] Upstream primer LOP-F2: 5'CACTCGTCCGGACAACGGTGGTGGAGGTAGTG 3'
[0067] Downstream primer LOP-R2:5'CC CTCGAG GCTACAGTTGGCGGTACGA 3' (the underlined part is the XhoI restriction site)
[0068] Using artificially synthesized Lpp-OmpA gene and PETase gene as templates, the target genes Lpp-OmpA and PETase were obtained by PCR amplification. The PCR reaction parameters were: pre-den...
Embodiment 3
[0089] Example 3: Construction and application of recombinant Escherichia coli (BL21 / pET22bBP) displaying PET decomposing enzyme on the cell surface
[0090] Chemically synthesize the BrkA gene shown in SEQ ID No.4 as the anchor protein, and chemically synthesize the PETase gene shown in SEQ ID No.1;
[0091] Primers were designed using the BrkA gene sequence and the PETase gene sequence as templates.
[0092] Upstream primer BrkA1 5'G GAATTC CATATGTACTTGGATCGTTTTCGGCA3' (the underlined part is the NdeI restriction site)
[0093] Downstream primer BrkA2 5'ATAAGGATTGGTTTGCCCGGCGTCCTGAGCATGT3'
[0094] Upstream primer BrkA3 5'ATGCTCAGGACGCCGGGCAAACCAATCCTTATGCC3'
[0095] Downstream primer BrkA4 5'GAGATATTCCGGCGCTACAGTTGGCGGTACGA3'
[0096] Upstream primer BrkA5 5'GCCAACTGTAGCGCCGGAATATCTCTTAGTGTT3'
[0097] Downstream primer BrkA6 5'CCG CTCGAG TCAAAACGAGTACCGATAGCC 3' (the underlined part is the XhoI restriction site)
[0098] The artificially synthesized BrkA gene and...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com