Method for detecting phalaenopsis amabilis mutants
A phalaenopsis and mutant technology, applied in biochemical equipment and methods, microbial determination/inspection, DNA/RNA fragments, etc., can solve problems such as inability to separate samples, poor experimental repeatability and stability, and save production costs. Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
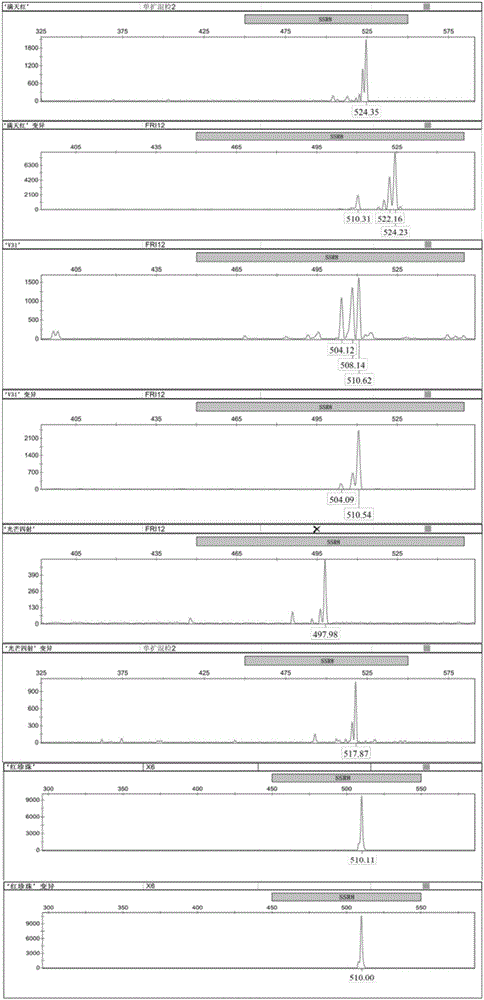
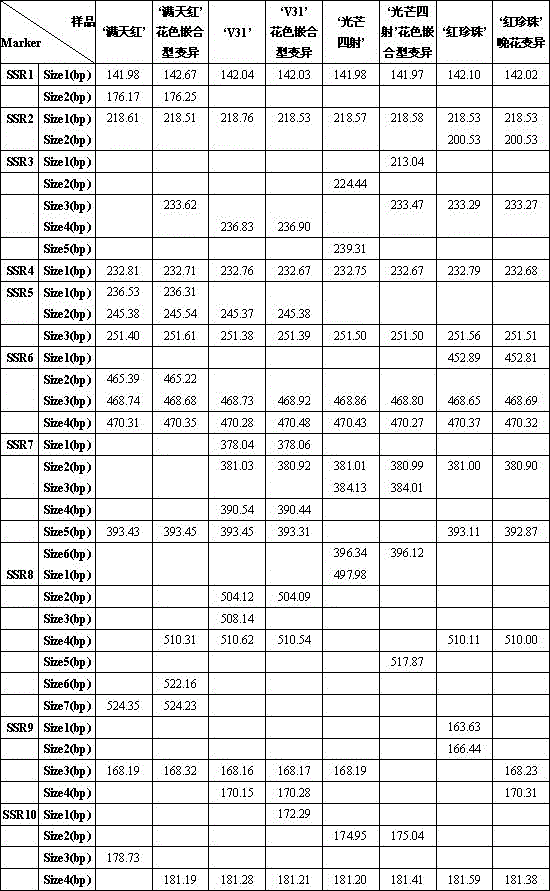
[0018] (1) Extraction of total DNA: Take 0.2 g of the root tip tissues of four Phalaenopsis 'Mantianhong', 'Raining Radiance', V31 and 'Red Pearl' respectively, and follow the conventional extraction methods or conventional reagents for total DNA in this field The total DNA of four kinds of Phalaenopsis 'Mantianhong', 'Radiance', V31 and 'Red Pearl' were extracted from the box.
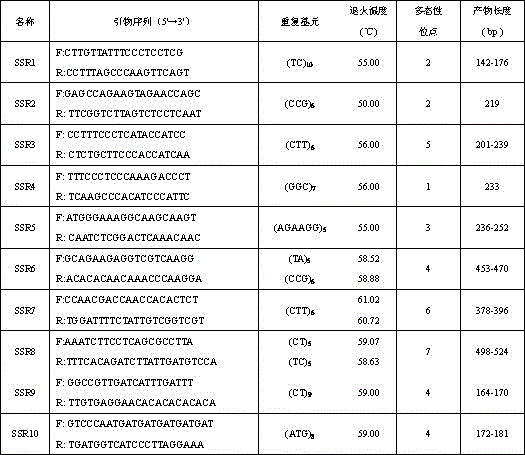
[0019] (2) PCR reaction: perform PCR with ten pairs of primers including SSR1, SSR2, SSR3, SSR4, SSR5, SSR6, SSR7, SSR8, SSR9 and SSR10.
[0020] The PCR reaction system was 6.0 µL of 2.5×Buffer V, 1.0 µL of upstream and downstream primers (5 µM), Taq enzyme (5U·µL -1 ) 0.1 µL, DNA 1.0 µL, ddH 2 O fill up to 15 µL;
[0021] The reaction program was 95 °C for 5 min, 35 cycles of 94 °C for 30 sec, 58 °C for 35 sec, 72 °C for 35 sec, and 60 °C for 30 min.
[0022] The sequences of ten pairs of primers for SSR1, SSR2, SSR3, SSR4, SSR5, SSR6, SSR7, SSR8, SSR9 and SSR10 are shown in Table 1.
[0023] Ta...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com