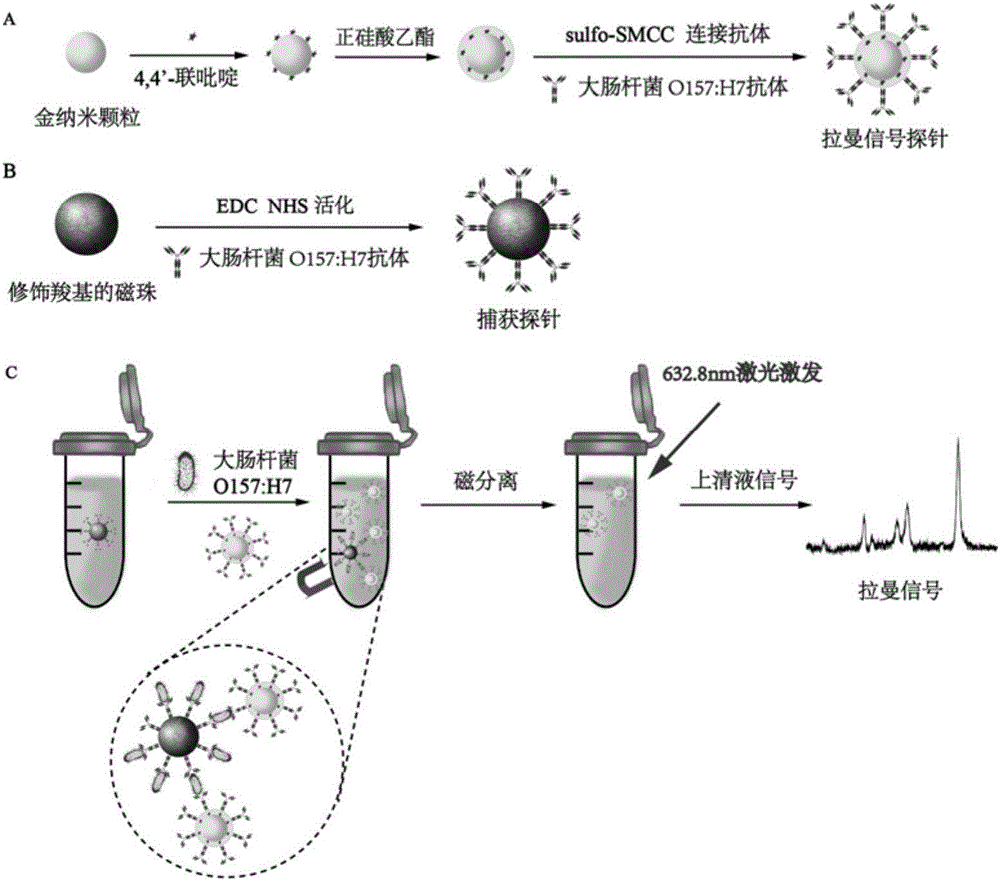
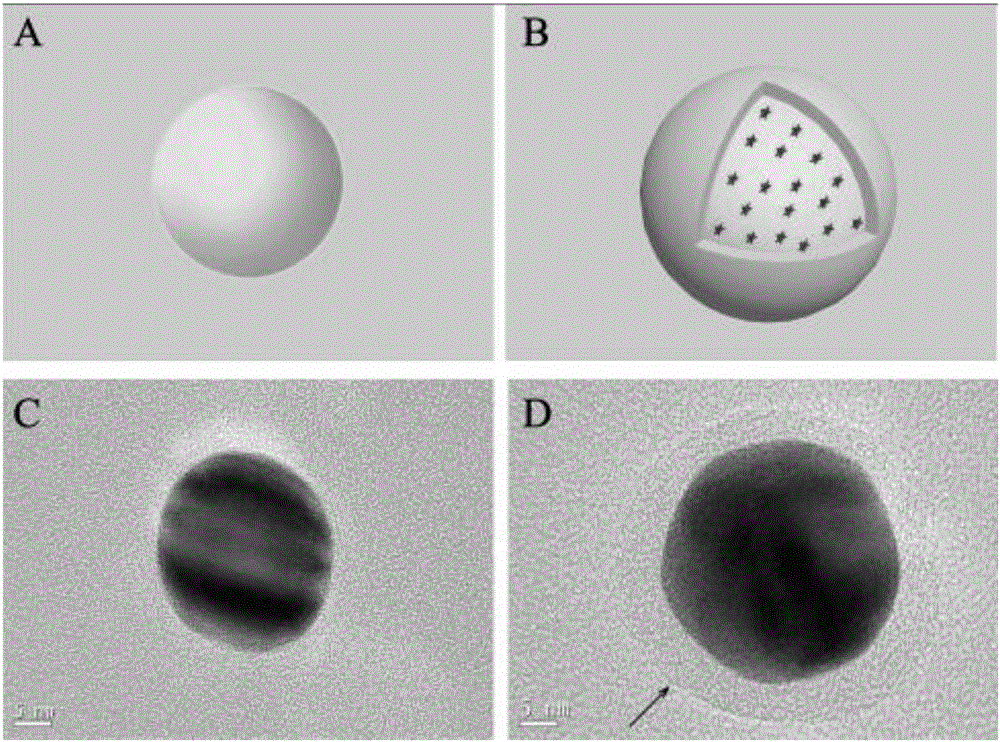
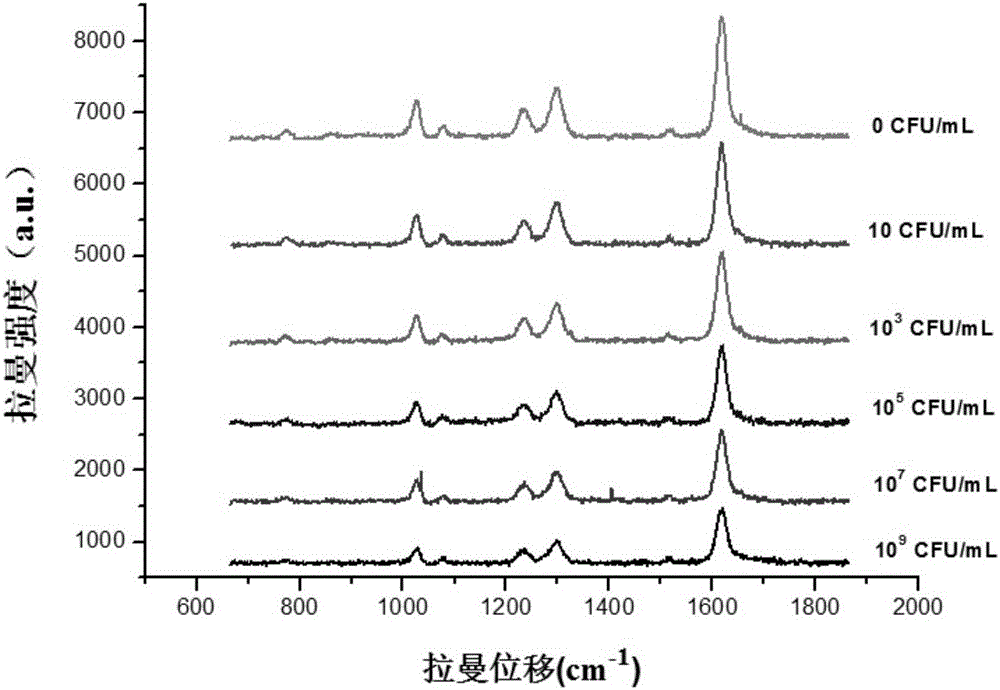
Novel SERS substrate-based method for quantitatively testing pathogenic bacteria
A technology for quantitative detection and pathogenic bacteria, which is applied in measuring devices, instruments, material analysis by optical means, etc., can solve the problems of low detection sensitivity, cumbersome and time-consuming process, and long growth time, so as to achieve high sensitivity and improved Capture efficiency and specificity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0053] (1), preparation of gold nanoparticles
[0054] Under constant stirring, 100 mL of 1 mM chloroauric acid (HAuCl 4 ) solution was heated to boiling, and then 6 mL of 38.8 mM trisodium citrate aqueous solution was added. At this time, the color change of the solution is: light yellow-colorless-black-purple-dark red, wait for the solution to turn dark red and continue to heat and reflux for 15-20min. Finally cooled to normal temperature, the preparation obtains the gold nanoparticle of 30nm (such as figure 2 C), figure 2 A is gold nanoparticles obtained under ideal conditions.
[0055] (2), preparation of silica-coated gold nanoparticles
[0056] Add 1mL gold nanoparticles (concentration is about 1mmol / L) to six 1.5mL EP tubes respectively, then add 30μL 0.01mM 4,4′-bipyridine solution in turn, mix well and react for 10min, centrifuge at 8000rpm for 10min, Remove the supernatant and resuspend with 1 mL triple distilled water. Then all the colloids in the 6 EP tubes...
Embodiment 2
[0069] Take 1 mL of the prepared gold nanoparticle solution, put it into an EP tube, add 30 μL of 0.01 mM 4,4′-bipyridine solution, react at room temperature for 10 min, centrifuge at 8000 rpm (10 min), remove the supernatant, and wash with BB buffer Resuspend, add 5μL 0.5mg / mL rabbit anti-E.coli O157:H7, incubate for 2h, centrifuge (8000rpm, 10min) to remove the supernatant, resuspend in BB buffer, then add 30μL 2.5% BSA to block unbound site, centrifuged (8000rpm, 10min) after 1h to remove the supernatant, and resuspended with PBS buffer to prepare bare gold signal probes. Place it with the signal probe in embodiment 1 step (3) for the same time, and measure the Raman signal intensity (such as Figure 5 ). from Figure 5 According to the report, preliminary studies have shown that this new type of SERS substrate has the following advantages: 1) the silicon dioxide silicon layer can ensure that gold nanoparticles are not disturbed by external chemical conditions; The inner...
Embodiment 3
[0071] Take the water body sample of the Guangzhou section of the Pearl River, filter, centrifuge, and dilute 40 times with triple distilled water to eliminate the influence of the matrix (the concentration of E.coli O157:H7 in the water body dilution of the known concentration is 10 2 、10 4 、10 6 、10 8 CFU / mL). According to the Raman spectrum signal intensity obtained in the actual water sample, from Figure 4 Read the corresponding concentration of E.coliO157:H7 from the standard curve in , and multiply it by the corresponding dilution factor to get the actual concentration of E.coli O157:H7 in the sample to be tested. The concentration of the actual water sample obtained according to the linear equation in Example 1 is shown in Table 1 below.
[0072] E.coli O157:H7 concentration and the E.coliO157:H7 concentration measured by the method of this embodiment in the actual water body sample of table 1
[0073] Known Concentration (CFU / mL) Measured concentratio...
PUM
| Property | Measurement | Unit |
|---|---|---|
| particle size | aaaaa | aaaaa |
| particle diameter | aaaaa | aaaaa |
| wavelength | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com