Method and system for predicting amino acid mutation
A technology of amino acids and algorithms, applied in the field of bioinformatics, can solve the problems of blindness and cost of biological experiments, achieve the effect of solving blindness and high cost, improving accuracy and effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
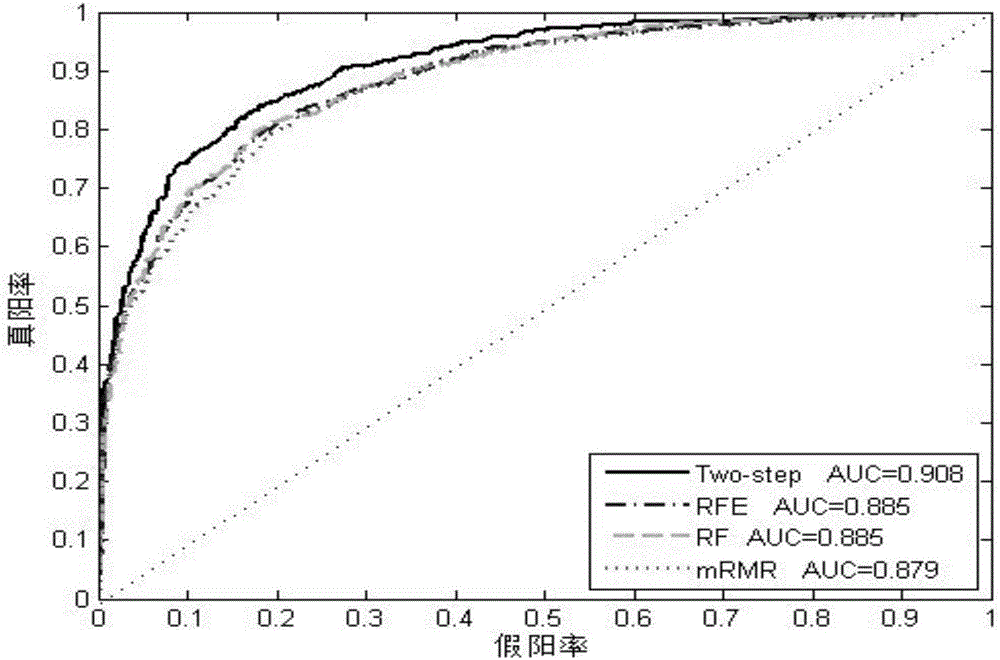
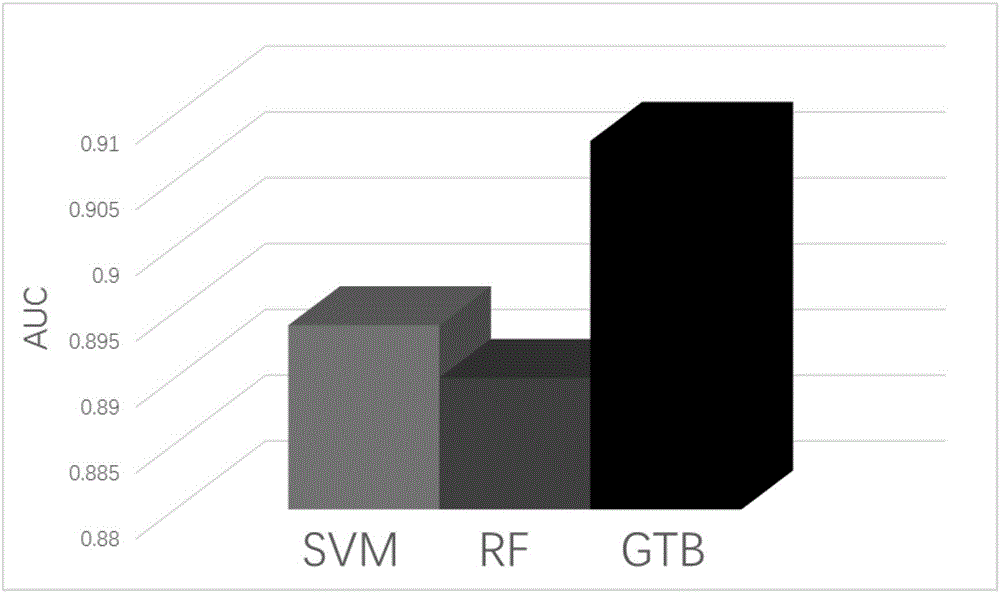
Image
Examples
Embodiment 1
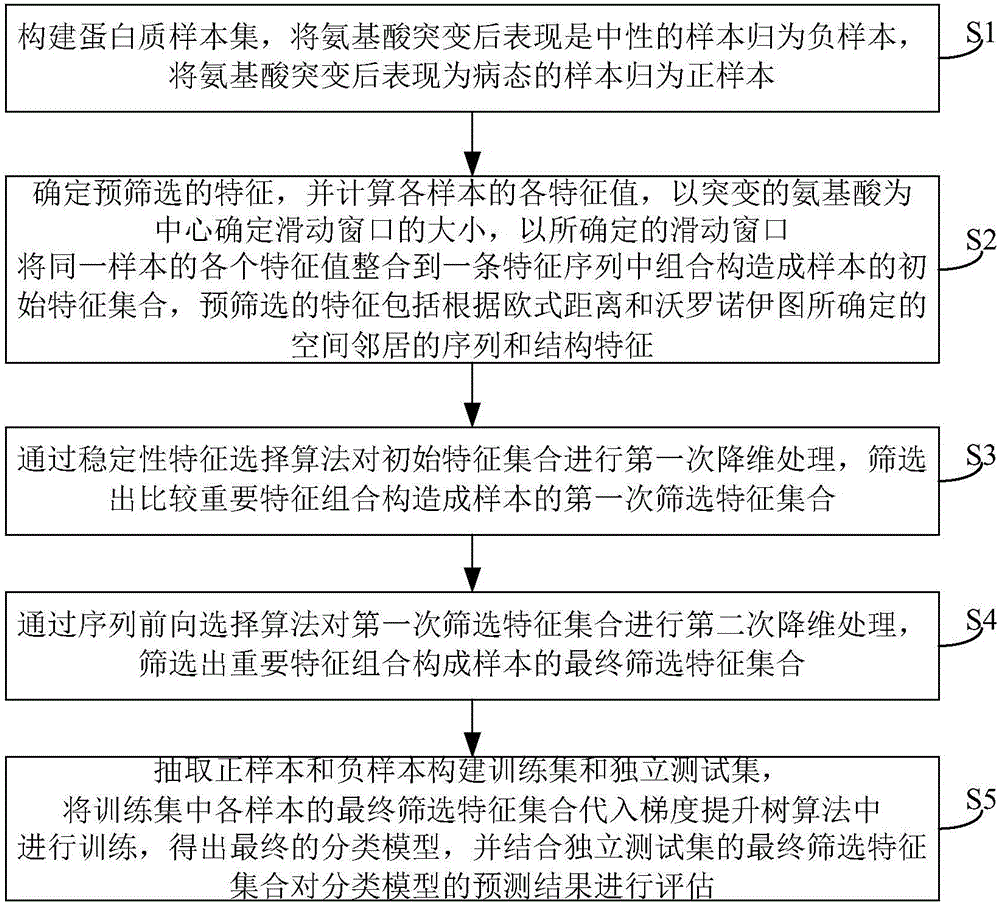
[0028] This embodiment discloses a method for predicting amino acid mutations, such as figure 1 shown, including:
[0029] Step S1, constructing a protein sample set, classifying the samples that are neutral after amino acid mutation as negative samples, and classifying the samples that are pathological after amino acid mutation as positive samples.
[0030] In this step, the data of negative samples can be extracted from the Ensemble human variation database. The data of the positive samples can be extracted from the UniProt human sequence variations database.
[0031] Preferably, in the process of constructing the protein sample set, repetitive data with a protein sequence similarity greater than 0.4 can be eliminated first.
[0032] Step S2, determine the characteristics of the pre-screening, and calculate the eigenvalues of each sample, determine the size of the sliding window centered on the mutated amino acid, and integrate the eigenvalues of the same sample into a...
Embodiment 2
[0060] Corresponding to the above method embodiments, this embodiment discloses a system for predicting amino acid mutations, including the following first to fifth processing modules. The functions of each module are described as follows:
[0061] The first processing module is used to construct a protein sample set, classify the samples that are neutral after amino acid mutations as negative samples, and classify the samples that are pathological after amino acid mutations as positive samples. Among them, the data of the negative sample can be extracted from the Ensemblehumanvariation database; the data of the positive sample can be extracted from the UniProt human sequence variations database. Preferably, the first processing module is also used to eliminate repetitive data with a protein sequence similarity greater than 0.4 during the process of constructing the protein sample set, thereby increasing the gold content of the data samples.
[0062] The second processing mod...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com