Method of using phospholipase C to extracellularly express intracellular protein
A phospholipase and protein technology, which is applied in the direction of microorganism-based methods, biochemical equipment and methods, and the use of carriers to introduce foreign genetic materials, etc., can solve the problems of high cost and cumbersome extraction process, and achieve the improvement of cell membrane permeability and wide application value effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0045] Example 1: Co-expression of L. monocytogenes phospholipase C and sucrose phosphorylase
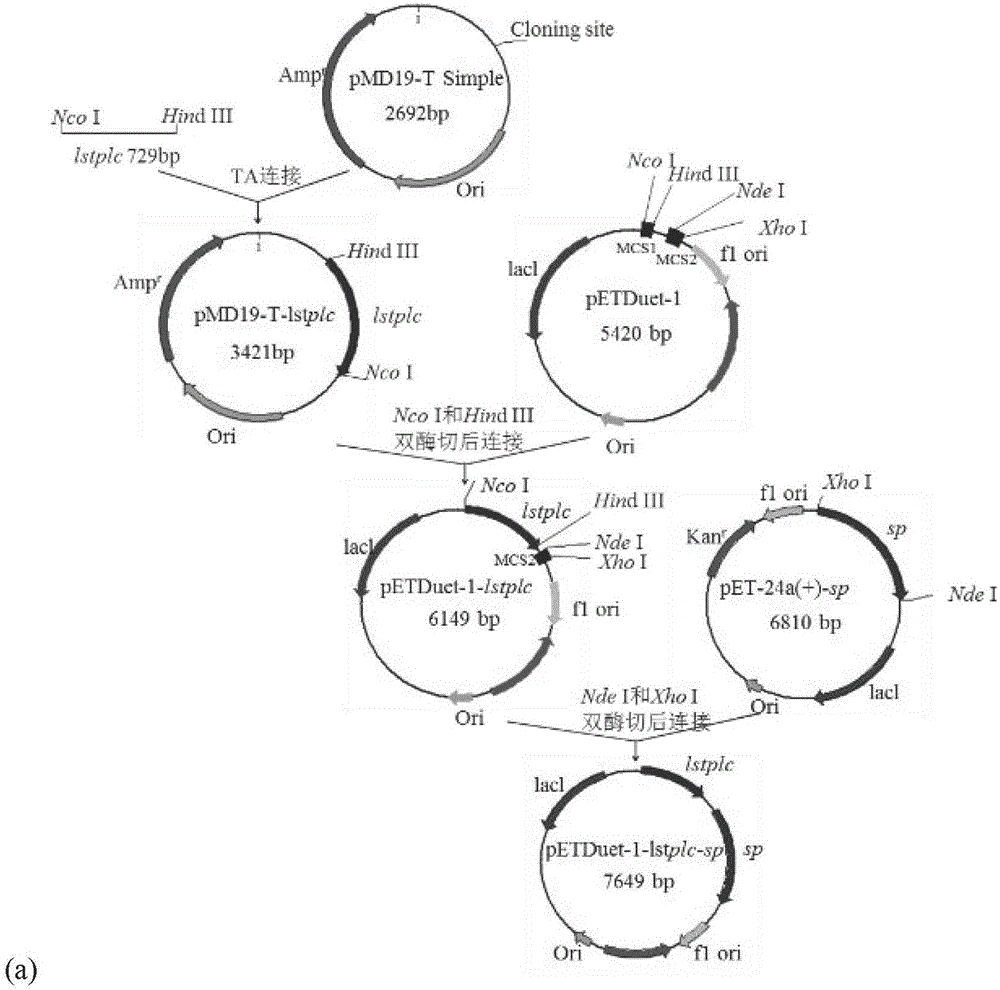
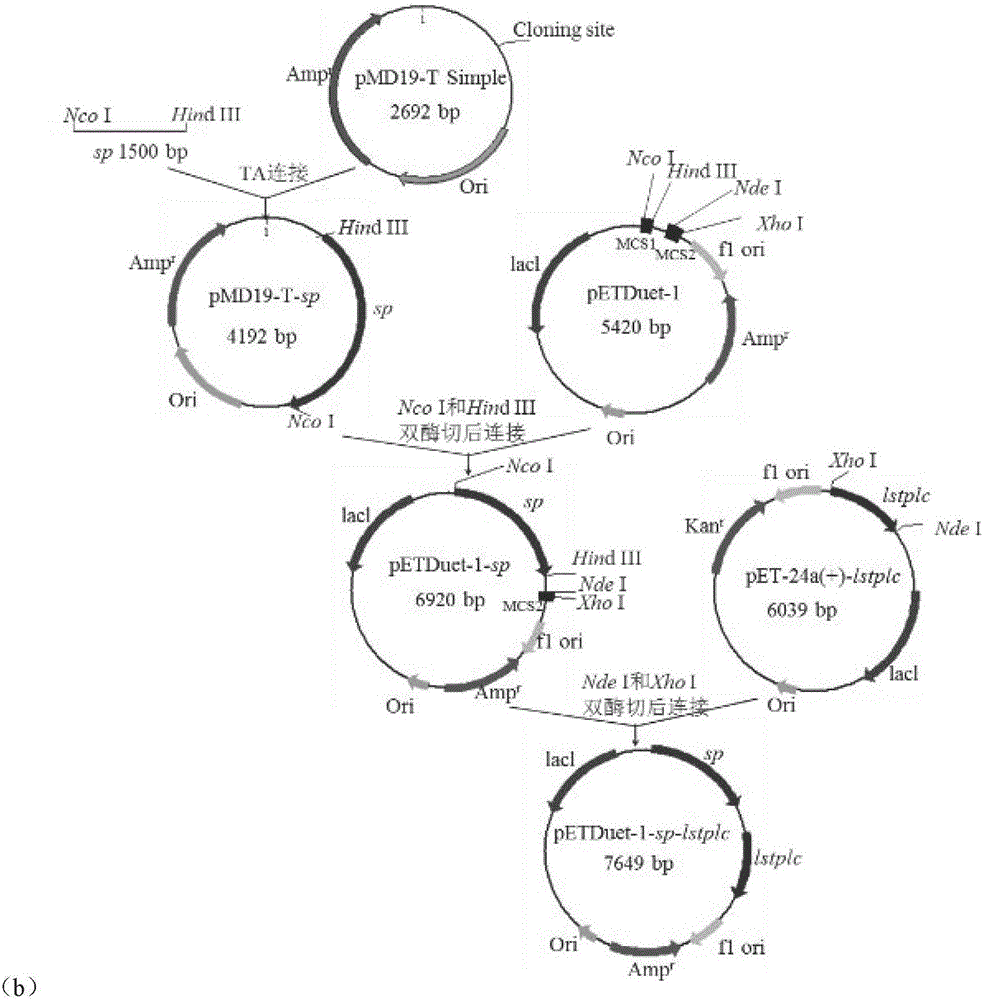
[0046] 1. Construction of recombinant bacteria: respectively with the expression plasmid pET-24a(+)-lstplc containing the L.monocytogenes phospholipase C gene (amino acid sequence shown in SEQID NO.1) and the sucrose phosphorylase gene preserved in the laboratory The expression plasmid pET-24a(+)-sp was used as a template, and PCR amplification obtained phospholipase C gene lstplc and sucrose phosphorylase gene sp containing Nco I and Hind III restriction sites, and then according to figure 1 (a) and (b) Plasmid construction process Insert L.monocytogenes phospholipase C and sucrose phosphorylase genes into the multiple cloning site MCS1 or MCS2 of the pETDuet-1 (purchased from Novagen) expression vector, respectively, and transform the ligated product The cloning host E.coli JM109 was coated with LB-Amp solid medium, cultivated in a 37°C incubator for 8 hours, picked a single colon...
Embodiment 2
[0049] Example 2: Co-expression of C. perfringens phospholipase C and sucrose phosphorylase
[0050] 1. Construction of recombinant bacteria: respectively with the expression plasmid pET-24a(+)-cpplc containing the C. perfringens phospholipase C gene (amino acid sequence shown in SEQ ID NO.2) and the sucrose phosphorylase gene preserved in the laboratory The expression plasmid pET-24a(+)-sp was used as a template, and PCR amplification obtained phospholipase C gene cpplc and sucrose phosphorylase gene sp containing Nco I and Hind III restriction sites, and then referred to figure 1 (a) and (b) Plasmid construction process Insert the C. perfringens phospholipase C and sucrose phosphorylase genes into the multiple cloning site MCS1 or MCS2 of the pETDuet-1 expression vector, and transform the ligated product into the cloning host E.coli JM109 Coat LB-Amp solid medium, culture in 37°C incubator for 8 hours, pick a single colony, and after 37°C culture in LB-Amp liquid medium for ...
Embodiment 3
[0052] Example 3: Co-expression of B. cereus phospholipase C and sucrose phosphorylase
[0053] 1. Construction of recombinant bacteria: respectively with expression plasmid pET-24a(+)-bcplc containing B. cereus phospholipase C gene (amino acid sequence shown in SEQ ID NO.3) and laboratory preservation containing sucrose phosphorylase The expression plasmid pET-24a(+)-sp of the gene was used as a template, and PCR amplification obtained phospholipase C gene bcplc and sucrose phosphorylase gene sp containing Nco I and Hind III restriction sites, and then referred to figure 1 (a) and (b) Plasmid construction process Insert the B. cereus phospholipase C and sucrose phosphorylase genes into the multiple cloning site MCS1 or MCS2 of the pETDuet-1 expression vector, and transform the ligated product into the cloning host E.coli JM109 Coat LB-Amp solid medium, culture in 37°C incubator for 8 hours, pick a single colony, and after 37°C culture in LB-Amp liquid medium for 8 hours, coll...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com