High-sensitivity gene mutation detection method and kit used in same
A detection kit and high-sensitivity technology, applied in biochemical equipment and methods, microbial measurement/inspection, etc., can solve the problems of long detection cycle, inaccurate detection results, low sensitivity, etc., and achieve low detection cost and convenient design Primer, high sensitivity effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0066] 1), the design method of LNA-modified ARMS-like primers:
[0067] The sequence (part) of the wild-type / mutant type MINA plasmid is as follows:
[0068] Mutation type (C / T)
[0069] aaagattttgatcagaaaagggcaacgattcagtttcaccaacctcagagatttaaggatgagctttggaggatccaggagaagctggaatgttactttggctccttggttggctcgaatgtgtacataactcccgcaggatctcagggcctgccgccccattatgatgatgtcgaggttttcatcctgcagctggagggagagaaacactggcgcctctaccaccccactgtgcccctggcacgagagtacagcgtggaggccgaggaaaggatcggcaggccggtgcatgagtttatgctgaagccgggtgatttgttgtactttcccagaggaaccatt C(T) atcaagcggacactcctgcggggctggcccactcgactcacgtgaccatcagcacctaccagaacaattcatggggagatttccttttggataccatctcggggcttgtatttgatactgcaaaggaagacgtggagttacggaccggcataccccggcagctgctcctgcaggtggaatccacaactgttgctacaagacgattaagtggcttcctgaggacacttgcagaccggctggagggcaccaaagaactgctttcctcagacatgaagaaggattttattatgcacagactccccccttactctgc。
[0070] The design rule of the LNA-modified ARMS-like primer is as follows: a base at the 3' end is modified to lock nucleic acid, the pr...
Embodiment 2
[0099] Example 2. Detection of mutations in exon 15 of the B-raf gene
[0100] The wild-type / mutant B-raf sequence (part) is as follows:
[0101] Mutation type (T / A)
[0102] aatcattgttttagacatacttattgactctaagaggaaagatgaagtactatgttttaaagaatattatattacagaattatagaaattagatctcttacctaaactcttcataatgcttgctctgataggaaaatgagatctactgttttcctttacttactacacctcagatatatttcttcatgaagacctcacagtaaaaataggtgattttggtctagctacagT(A)gaaatctcgatggagtgggtcccatcagtttgaacagttgtctggatccattttgtggatggtaagaattgaggctatttttccactgattaaatttttggccctgagatgctgctgagttactagaaagtcattgaaggtctcaactatagtattttcatagttcccagt。
[0103] Therefore, the designed LNA-modified ARMS-like primer is: SEQ ID NO: 4, and another conventional primer is SEQ ID NO: 5.
[0104] Numbering Primer sequence (5'-3') SEQ ID NO:4 GGTGATTTTGGTCTAGCTACAGA SEQ ID NO:5 CTGATGGGACCCACTCCATCGA
[0105] The rest are equal to Example 1.
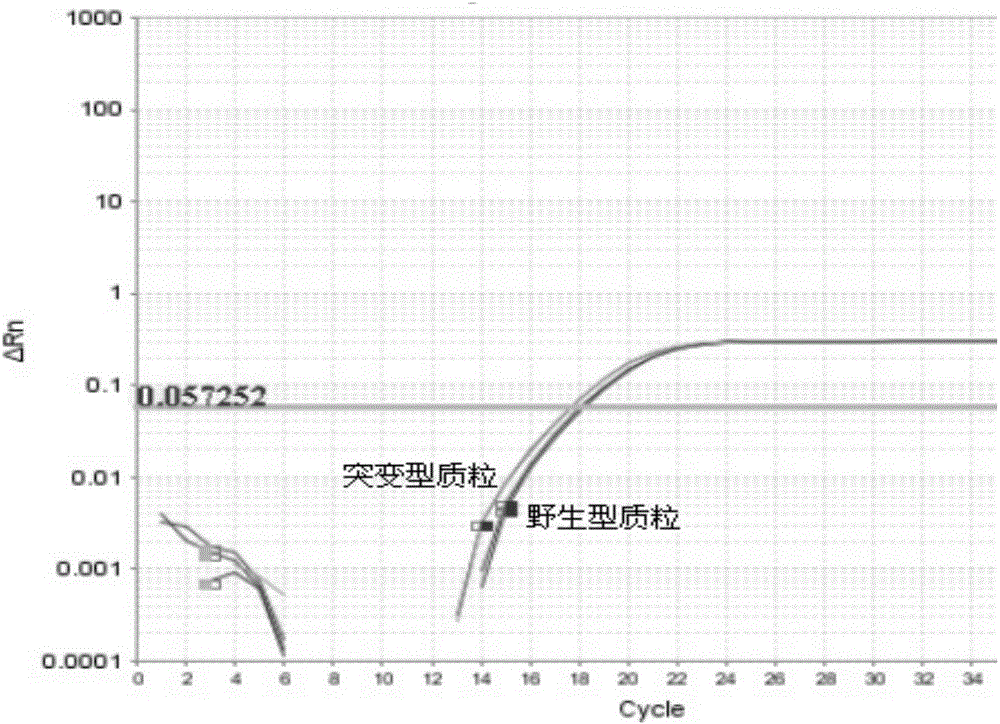
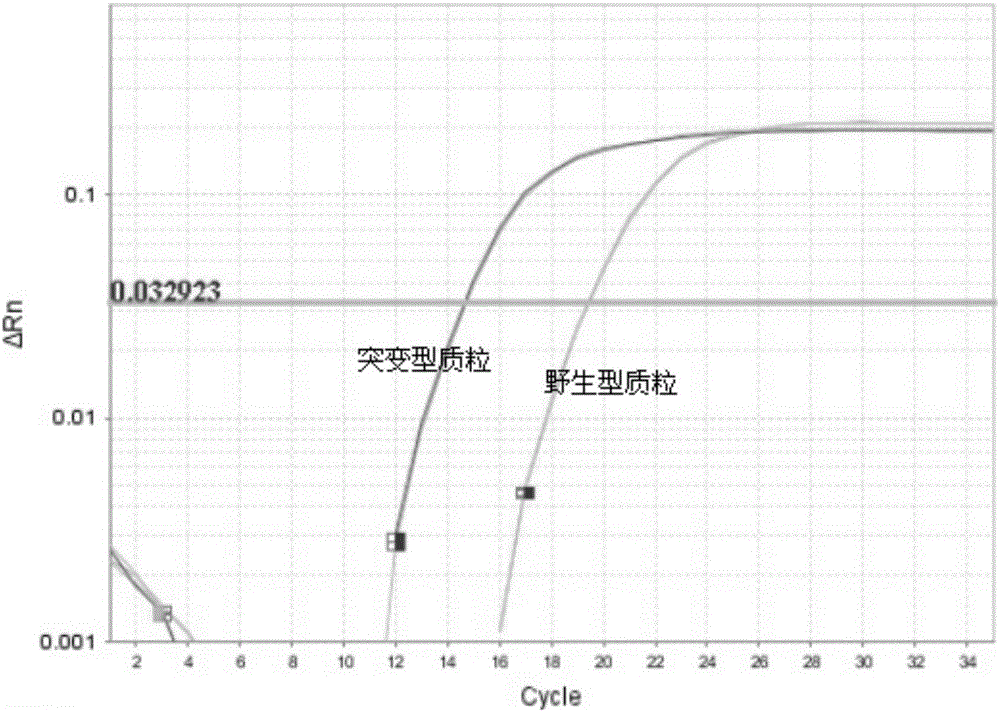
[0106] The result is as Figure 6 shown, according to Figure 6, we know that the Ct v...
Embodiment 3
[0107] Example 3. Detection of mutations in the 12th codon of exon 2 of the K-ras gene
[0108] The wild-type / mutant K-ras sequence (part) is as follows:
[0109] Mutation type (G / A)
[0110] attgaattttgtaaggtattttgaaataatttttcatataaaggtgagtttgtattaaaaggtactggtggagtatttgatagtgtattaaccttatgtgtgacatgttctaatatagtcacattttcattatttttattataaggcctgctgaaaatgactgaatataaacttgtggtagttggagctgG(A)tggcgtaggcaagagtgccttgacgatacagctaattcagaatcattttgtggacgaatatgatccaacaatagaggtaaatcttgttttaatatgcatattactggtgcaggaccattctttgatacagataaaggtttctctgaccattttcatgagtacttattac。
[0111] Therefore, the designed LNA-modified ARMS-like primer is: SEQ ID NO:6, and another conventional primer is SEQ ID NO:7.
[0112] Numbering Primer sequence (5'-3') SEQ ID NO:6 ACTTGTGGTAGTTGGAGCTGA SEQ ID NO:7 TCGTCAAGGCACTCTTGCCTACG
[0113] The rest are equal to Example 1.
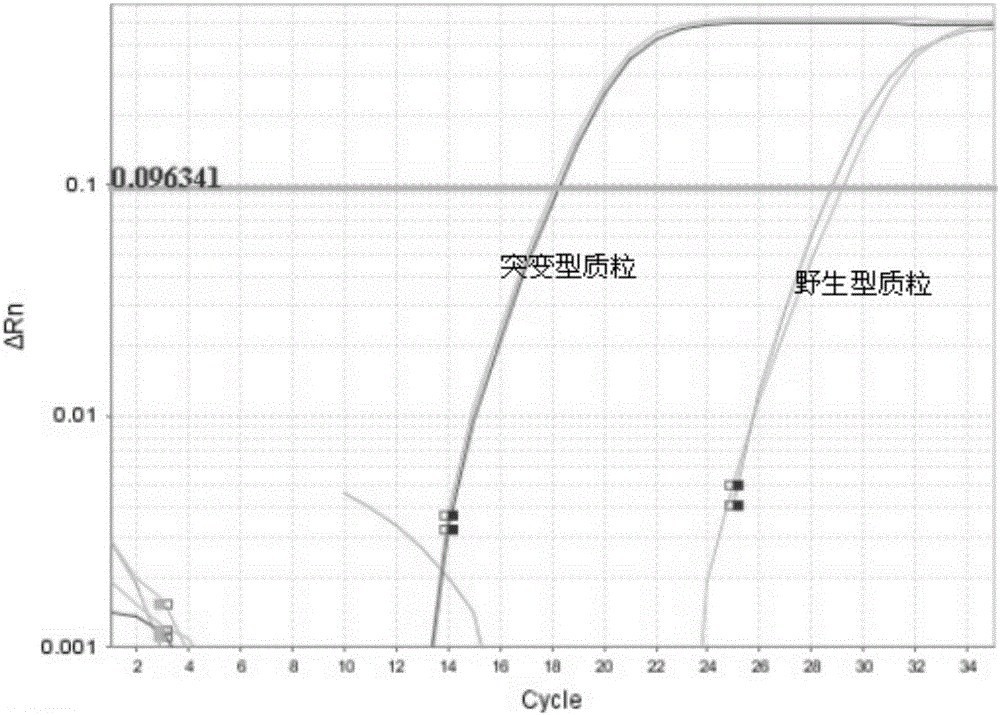
[0114] The result is as Figure 7 shown, according to Figure 7 , we know that the Ct value of the mutant sample i...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com