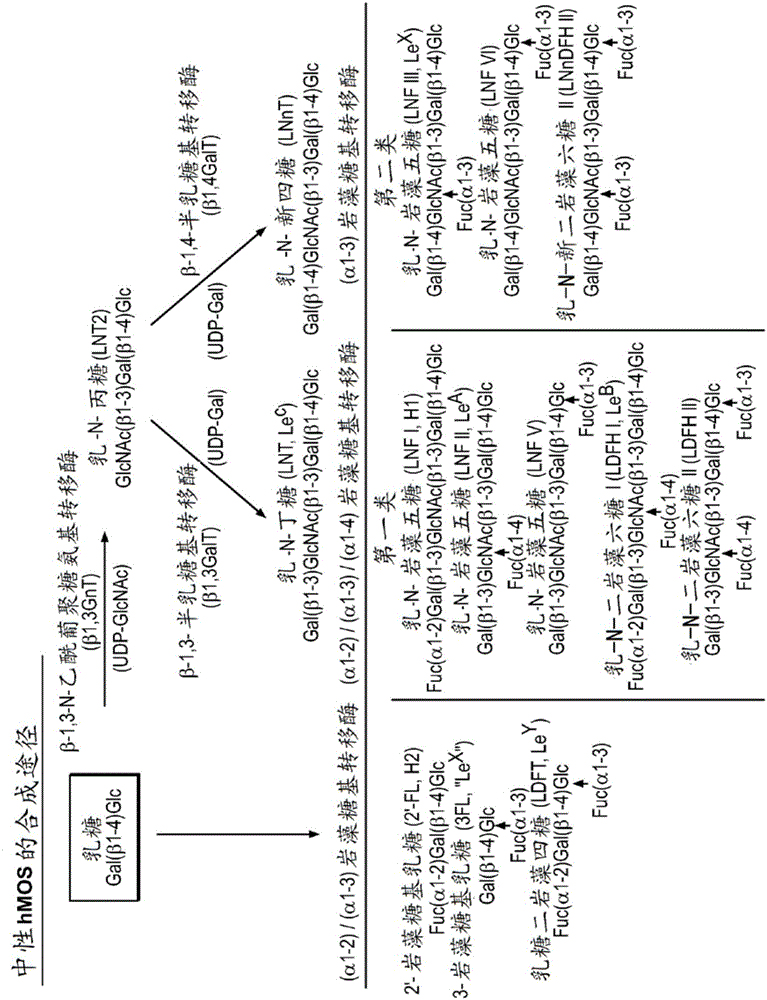
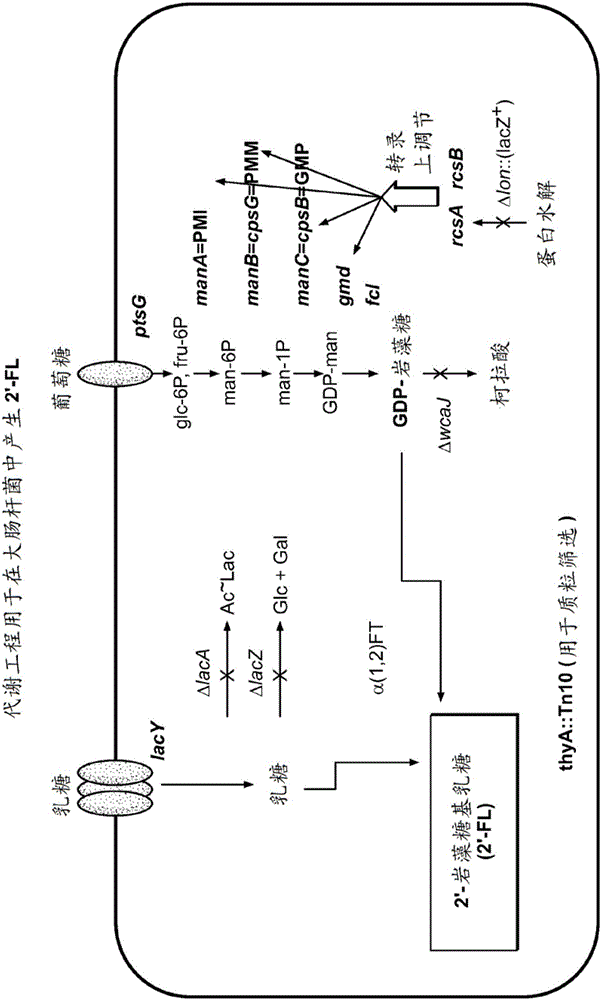
Alpha (1,2) fucosyltransferase syngenes for use in the production of fucosylated oligosaccharides
A technology of fucosyltransferase and fucosylation, which is applied in the directions of glycosyltransferase, transferase, esterification and saccharide, etc., can solve the problems of impure product, high cost, production limitation, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0154] Example 1: Identification of a novel α(1,2)-fucosyltransferase
[0155] In order to identify other novel α(1,2) fucosyltransferases, calibration operations were performed using CLCbio Main Workbench package version 6.9 to generate multiple sequence calibration queries (CLCbio, 10 Rogers Street #101, Cambridge, MA 02142, USA) using Four previously determined lactose-utilizing α(1,2) fucosyltransferase protein sequences: Helicobacter pylori futC (SEQ ID NO: 1), H. mustelae FutL (SEQ ID NO: 2), Bacteroides vulgaris futN (SEQ ID NO: 3) and E. coli 0126wbgL (SEQ ID NO: 4). Figure 3 shows the sequence alignment and percent sequence identity between four predetermined lactose-utilizing alpha(1,2) fucosyltransferase protein sequences. Iterative PSI-BLAST was performed, interrogated using FASTA-type multiple sequence alignments, and the NCBI PSI-BLAST program was run on a local copy of NCBI BLAST+ version 2.2.29. Specific streak matrix files (.pssm) were generated using PSI-...
Embodiment 2
[0162] Example 2: Verification of the new α(1,2)FT
[0163] To detect lactose-utilizing fucosyltransferase activity, fucosylated oligosaccharides (i.e., 2'-fucosyllactose ( 2'-FL)), and it contains cytoplasmic guanosine diphosphate-fucose and lactose pools. The production of fucosylated oligosaccharides indicated that the candidate enzyme-coding sequence functions as a lactose-utilizing alpha(1,2) fucosyltransferase. Of all the samples identified, 12 novel α(1,2) fucosyltransferases were further analyzed for their ability to produce 2′-fucosyltransferases: Prevotella melanogenes FutO, Clostridium Clostridium boltae+13FutP, Lachnospiraceae FutQFutP, Methanosphaerula palustris FutR, Tannerella sp. FutS, Bacteroides faecalis FutU, Butyvibrio FutV, Revobacteria FutW, parabacteroides johnsonii FutX, Akkermansia muciniphilia FutY, Salmonella enterica FutZ, Bacteroides FutZA.
[0164] The construct includes 12 variants of the novel α(1,2)FT with the following structure: EcoRI-T7...
Embodiment 3
[0173] Example 3: Characterization of cultures expressing novel α(1,2)FTs
[0174] Bacteria expressing the novel α(1,2)FTs FutO, FutQ and FutX were further characterized. Specifically, proliferation ratio and exogenous α(1,2) FT experiments were examined.
[0175] Expression plasmids containing the fucosyltransferases WbgL (plasmid pG204), FutN (plasmid pG217), and the novel α(1,2)FT FutO (plasmid pG393), FutQ (plasmid pG395), and FutX (pG401) were introduced into host bacteria strain. For example, the host strain utilized here has the following genotype: ΔampC::P trp B cI,Δ(1acI-lacZ)::FRT,P lacIq lac Y + , ΔwcaJ::FRT, thyA::Tn10, Δlon:(npt3, lacZ + ), ΔlacA
[0176] Bacterial cultures expressing each exogenous fucosyltransferase were induced by the addition of tryptophan in the presence of lactose (in order to induce expression of the exogenous fucosyltransferase). Growth of the cultures was monitored by recording spectrophotometer readings at A600 at the following...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com