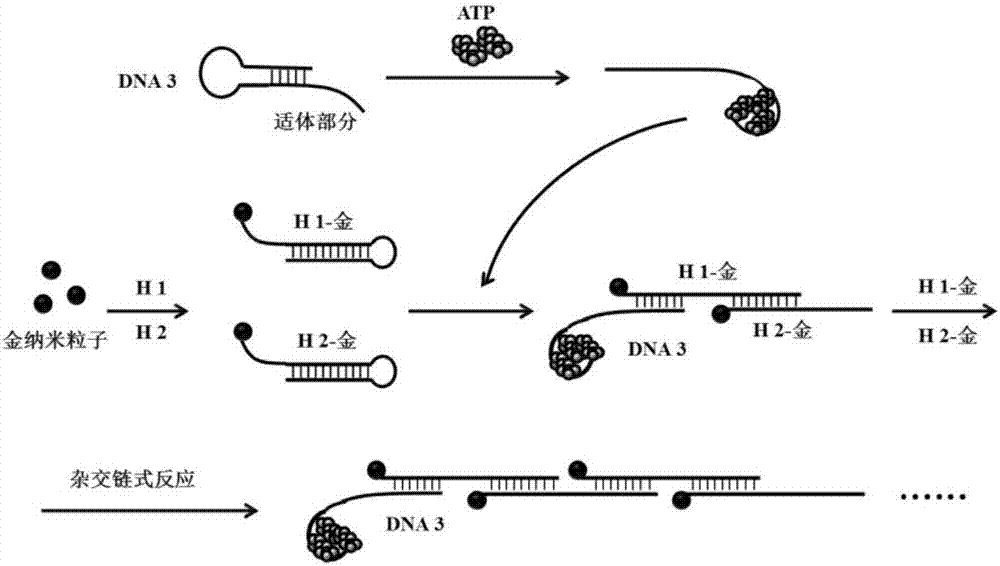
Method and probe for detecting miRNA (Ribonucleic Acid) and/or target molecule with aptamer
A nucleic acid aptamer and probe technology, applied in biochemical equipment and methods, DNA/RNA fragments, DNA preparation, etc., can solve problems such as high cost and complicated operation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0064] Embodiment 1: Detect the preparation of miRNA probe:
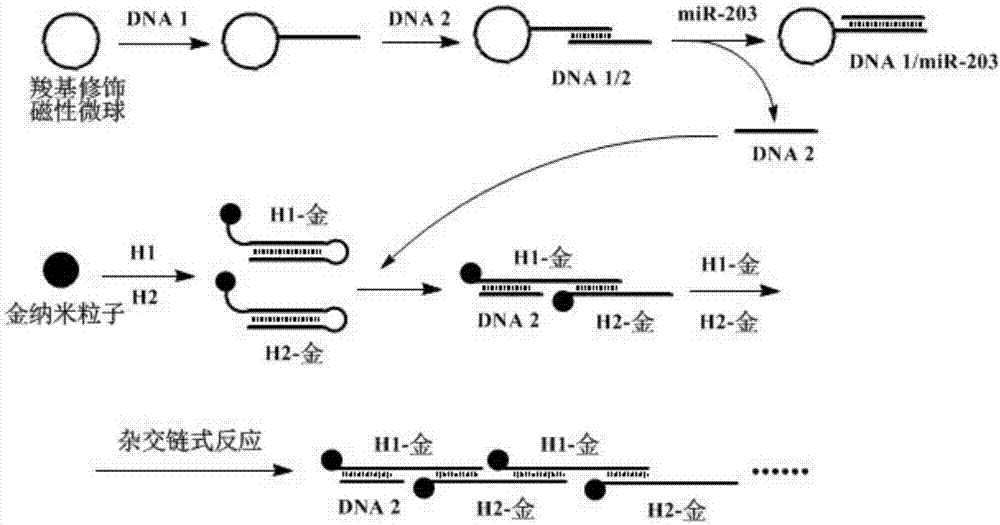
[0065] The probe includes: gold nanoparticle-H1 complex, gold nanoparticle-H2 complex and carboxyl magnetic microsphere-DNA1 / 2 complex; wherein: the sequences of H1 and H2 are as follows:
[0066] H1: 5'-SH-GCG ATT CCT AGG TTG AGC CCA GGG TTT TTT CACAGT CCC TGG GCTCAA CCT AGG-3'; (SEQ ID NO.1)
[0067] H2: 5'-CCC TGG GCT CAA CCT AGG AAT CGC TTT TTT CCT AGG TTGAGC CCA GGGACT GTG-SH-3'; (SEQ ID NO. 2).
[0068] DNA1 contains a sequence complementary and / or partially complementary to the miRNA to be detected; DNA1 is partially complementary to DNA2.
[0069] Wherein, the preparation method of gold nanoparticle-H1 composite, gold nanoparticle-H2 composite is:
[0070] (1) Preparation of gold nanoparticles with a diameter of 5nm: Add 0.6mL of 0.1M sodium borohydride on ice to 20mL of chloroauric acid (0.25mM) and trisodium citrate (0.25mM) mixture, and stir until the solution becomes Pink, reaction 2-5h. The gold nan...
Embodiment 2
[0074] Embodiment 2: the application of the probe of the present invention in detecting miR-203
[0075] 1. Test material:
[0076] The nucleotide sequence of miR-203 is as follows:
[0077] 5'-GUG AAA UGU UUA GGA CCA CUA G-3'.
[0078] The composition and preparation method of the probe are the same as in Example 1, in the probe:
[0079] The sequences of H1 and H2 are as follows:
[0080] H1: 5'-SH-GCG ATT CCT AGG TTG AGC CCA GGG TTT TTT CACAGT CCC TGG GCTCAA CCT AGG-3'; (SEQ ID NO.1)
[0081] H2: 5'-CCC TGG GCT CAA CCT AGG AAT CGC TTT TTT CCT AGG TTGAGC CCA GGGACT GTG-SH-3'; (SEQ ID NO.2);
[0082] The sequences of DNA1 and DNA2 are as follows:
[0083] DNA1: 5'-GGG C TAG TGG TCC TAA ACA TTT CAC-NH 2 -3'; (SEQ ID NO.3)
[0084] DNA2: 5'-GGA CCA CTA G CCC TGG GCT CAA CCT AGG AAT CGC-3'; (SEQ ID NO. 4).
[0085] 2. Detection method:
[0086] The specific method is as follows:
[0087] (1) Preparation of miR-203 working solution: Preparation of miR-203 working solution...
Embodiment 3
[0091] Example 3: Selectivity investigation of miR-203 detection method
[0092] miR-203, miR-21 (nucleotide sequence: 5'-UAG CUU AUC AGA CUG AUG UUG A-3') and miR-16 (nucleotide sequence: 5'-UAG CAG CAC GUA AAU AUU GGC G-3') solution Preparation: Sterilize the PBS buffer solution (pH 7.4) containing 1‰ (volume ratio) DEPC water, and use it to prepare 1.0×10 -10 M miR-203, 2.0×10 -9 MmiR-21 and 2.0×10 -9 M miR-16 solution.
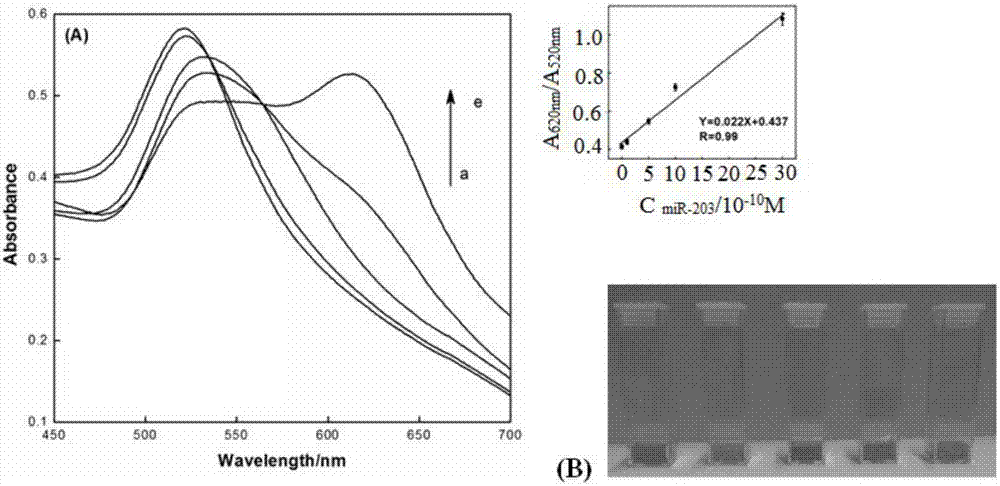
[0093] Mix 100 μL of carboxylated magnetic microsphere-DNA1 / 2 complex with 100 μL of miR-203 solution (1.0 × 10 -10 M) or miR-21 solution (1.0×10 -9 M) or miR-16 solution (1.0×10 -9 M), mix evenly, perform magnetic separation after reacting at 25°C for 2 hours, and absorb the supernatant, then add 100 μL gold nanoparticle-H1 complex and 100 μL gold nanoparticle-H2 complex to the supernatant, and incubate for 4 hours Finally, record the UV absorption peak ratio at 620nm and 520nm. Such as Figure 5 As shown, only the miR-203 solution caused signific...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com