Cas9 nuclease G915F and application thereof
A technology of nuclease and nuclease activity, which is applied in the direction of application, hydrolase, and the introduction of foreign genetic material using vectors, etc., to achieve the effect of precise editing
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
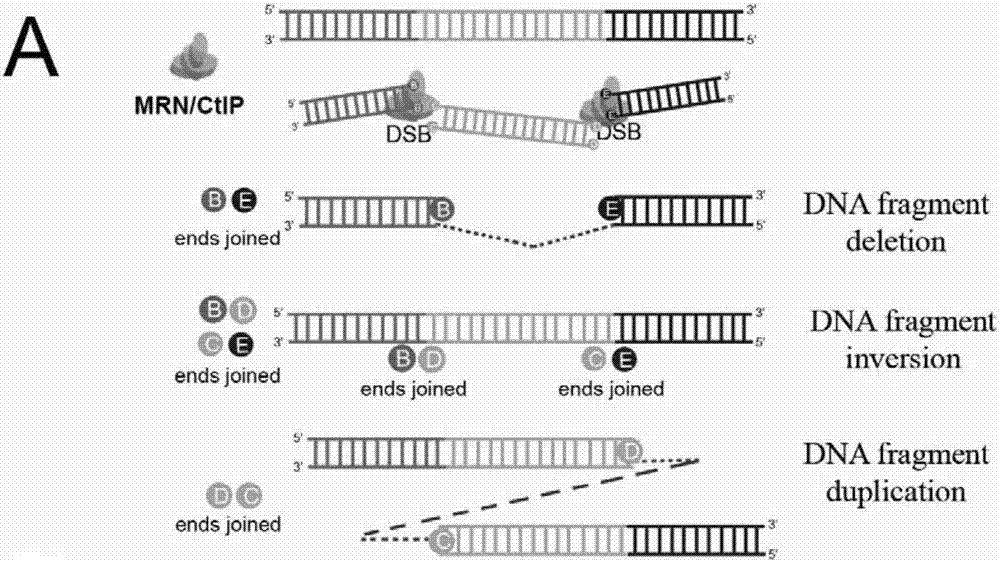
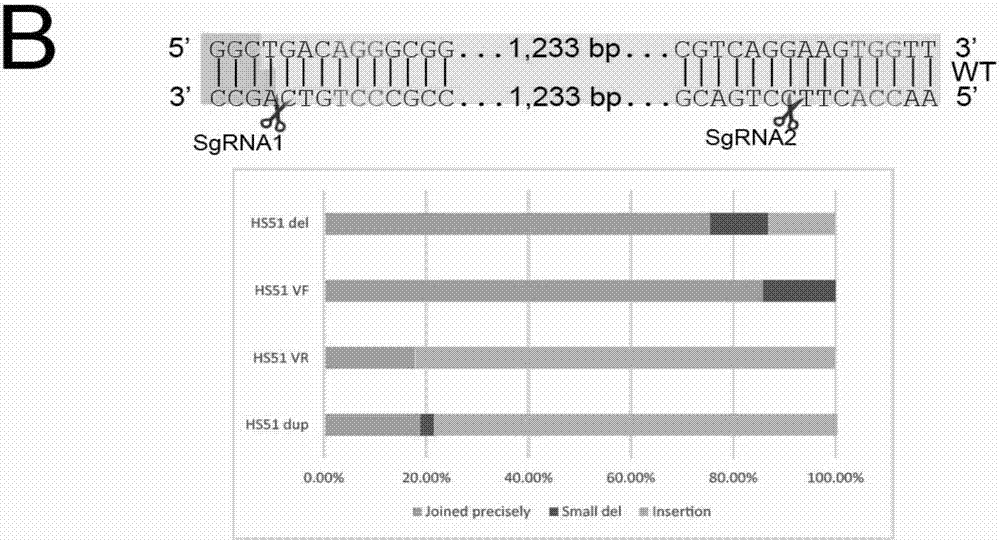
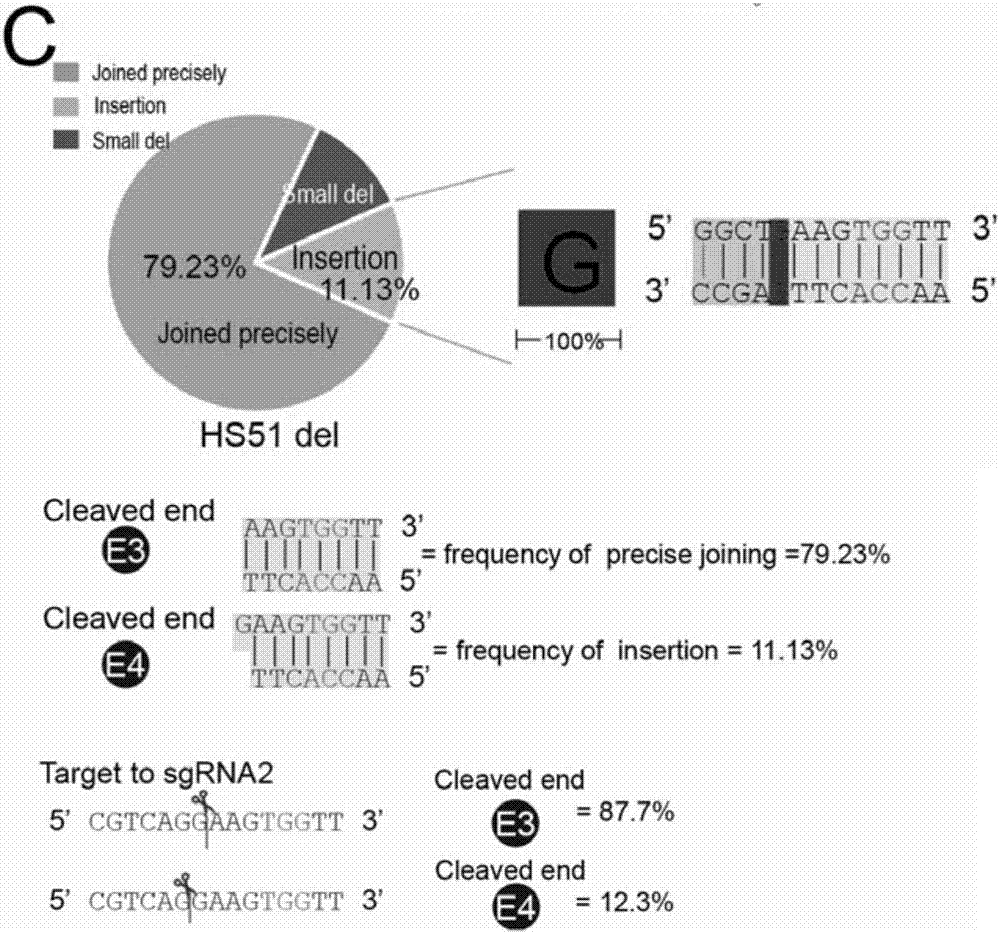
[0077] Example 1 Studying the Ligation of DNA Fragment Editing Adapters and Discovering a New Mechanism of Cas9 Cutting
[0078] For the HS51 site, construct the sgRNAs plasmid targeting the HS51 site:
[0079] (1) Purchase primers
[0080] Purchase forward and reverse deoxy oligonucleotides with 5' hanging ends "ACCG" and "AAAC" that can be complementary paired for the HS51 site and the sgRNAs targeting sequence respectively from Shanghai Sunny Biotechnology Co., Ltd.;
[0081] Targeting sequences of sgRNAs targeting the above HS51 site:
[0082] HS51 RE1 sgRNA1: GCCACACATCCAAGGCTGAC (SEQ ID NO.1)
[0083] HS51 RE1 sgRNA2: GAGATTTGGGGCGTCAGGAAG (SEQ ID NO.2)
[0084] (2) Obtain complementary paired double-stranded DNA with hanging ends
[0085] 1) use ddH 2 O Dissolve deoxyoligonucleotides to 100 μM and dilute to 20 μM;
[0086] 2) Add positive and negative deoxy oligonucleotides to the following reaction system:
[0087]
[0088]
[0089] Reaction conditions: 95...
Embodiment 2
[0153] Example 2 Mutation of SpCas9 to obtain a specific Cas9 with a changed cutting method to achieve precise DNA fragment editing
[0154] 1. Construction of Cas9 mutants
[0155] 1) Use NEB Mutagenesis Kit (Q5Site-Directed Mutagenesis Kit, #E0554S) to construct Cas9 mutants, first perform PCR amplification, and the reaction is as follows:
[0156]
[0157]
[0158]
[0159] 2) KLD (Kinase, Ligase&DpnI) treatment, the reaction is as follows:
[0160]
[0161] Reaction conditions: 10 minutes at room temperature
[0162] 3) All the reaction products in 2) were used for the transformation of competent bacteria Stbl3 (50 μl), and cultured on LB plates containing ampicillin antibiotic (Amp, 100 mg / L) overnight at 37° C. Single clones were picked, plasmids were extracted and sent for sequencing.
[0163] The amino acid sequence of SpCas9 (Cas9WT) is shown in SEQ ID NO.7, specifically:
[0164]
[0165]
[0166] The coding nucleotide sequence of SpCas9 (Cas9WT...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com