Aptamer of grass carp reovirus and derivative, screening method and application of aptamer
A nucleic acid aptamer, reovirus technology, applied in the field of molecular biology, can solve the problems of not being widely used, difficult screening process, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0065] Embodiment 1, the screening of the nucleic acid aptamer of grass carp reovirus
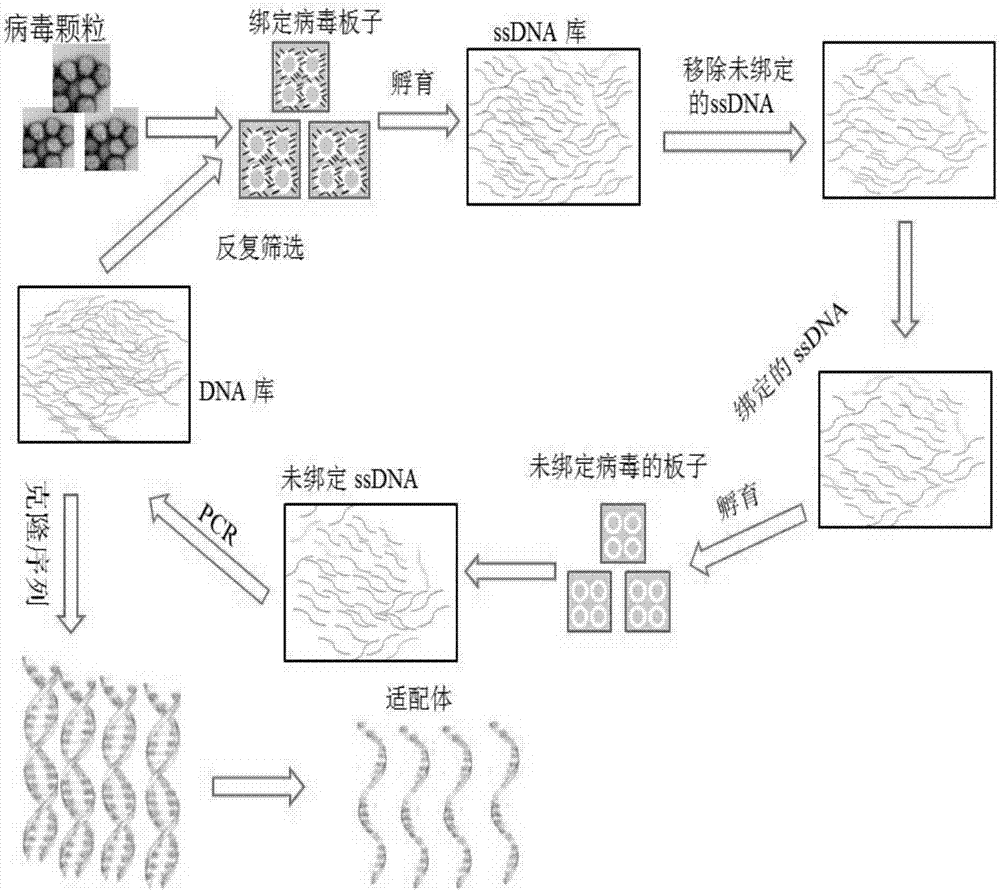
[0066] Use exponential enrichment ligand system evolution technology (SELEX technology) to screen the nucleic acid aptamers of grass carp reovirus, the SELEX technology schematic diagram can be found in figure 1 , the specific implementation method is as follows:
[0067] (1) Virus purification
[0068] GCRV was inoculated on FHM cells and cultured at 28°C until about 80% of the lesions appeared. After harvesting, it was frozen and thawed three times at –80°C, and the cell debris was removed by gradient centrifugation: 3000rpm / min, 15min; 5000rpm / min, 15min; 8000rpm / min, 20min; 10000rpm / min, 30min, to obtain GCRV virus liquid. At the same time, uninfected FHM cells were used as a control for the reverse screening of the screening process, and were also subjected to repeated freeze-thaw and gradient centrifugation under the same conditions to obtain FHM cell liquid, which was stored at –80...
Embodiment 2
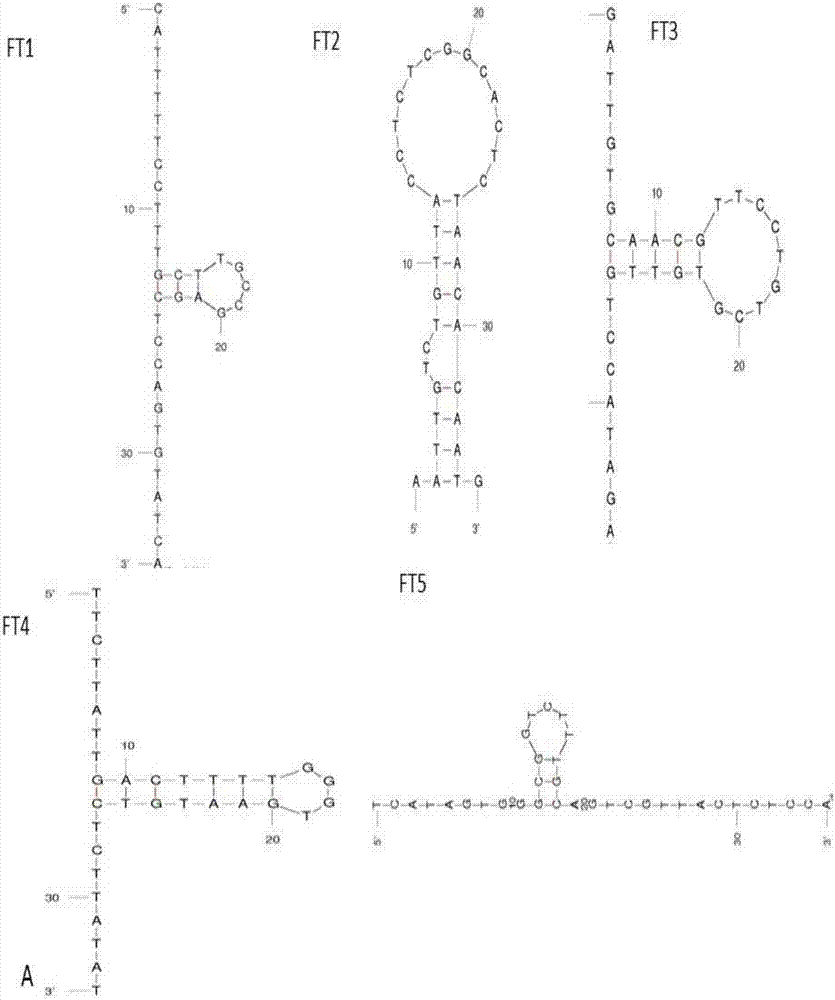
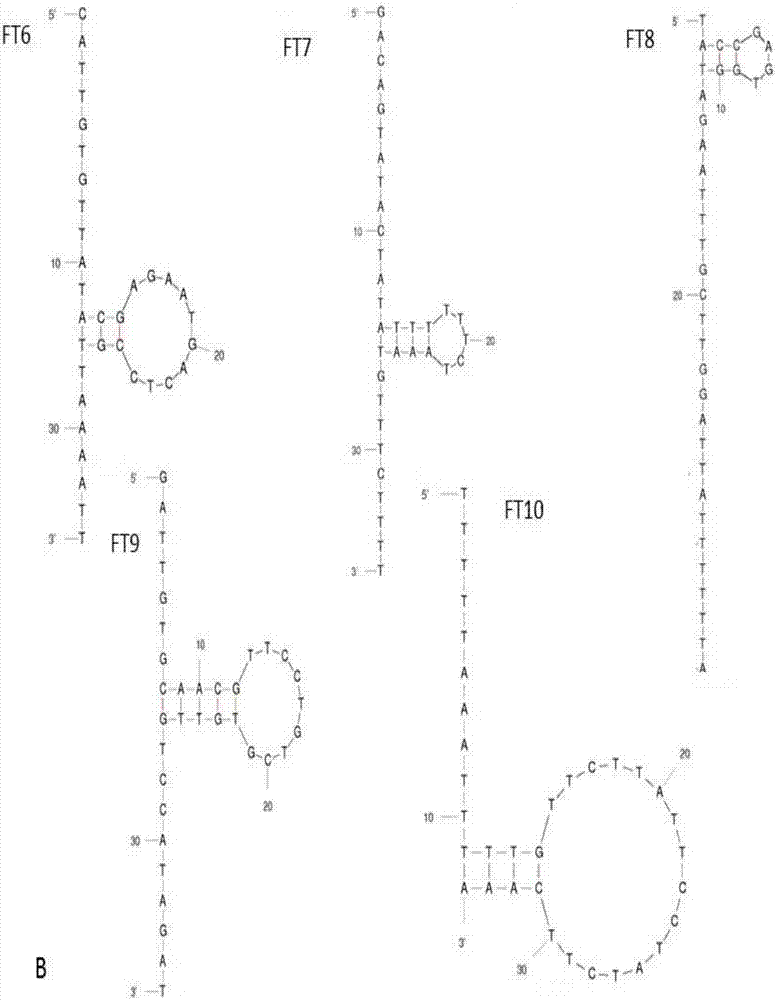
[0090] Example 2, Spatial Structure Prediction of Nucleic Aptamers
[0091] Predict the space structure of the 10 nucleic acid aptamers whose sequences were screened out above and whose frequency of repetition was determined: according to the energy principle of the MFOLD software, the minimum energy structure was selected as the most stable structure of the aptamer.
[0092] The result is as figure 2 As shown, all aptamers can form a stem-loop structure, which forms a stem-loop from about thirteen oligonucleotides to twenty-seven oligonucleotides, which may be the aptamer binding The key site of the target and play a role.
Embodiment 3
[0093] Example 3, the effect of nucleic acid aptamers on the viability of FHM cells
[0094] The FHM cells were seeded in a 96-well plate. When the cells grew to 80%, the aptamer FT1 was added at a concentration of 0 μM to 10 μM, cultured at 28°C for 48 hours, 20 μl / well of MTT solution was added, and cultured at 28°C for 4 hours. The ELISA plate reader reads at 490nm.
[0095] Experimental results such as image 3 As shown, after the nucleic acid aptamer FT1 was added to the cells at a concentration of 0uM to 10uM, the cell viability did not change significantly, indicating that the nucleic acid aptamer would not produce toxic side effects on the host cells, which is beneficial to the later preparation of drugs and the application of detection reagents.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com