Specifically-targeted swine IGFBP3 gene sgRNA targeting sequence and application
A kind of specific and sequence technology, applied in sgRNA guide sequence and application field, can solve the problems of mouse weight gain, expression level increase, content increase, etc., and achieve high safety and eliminate expression effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
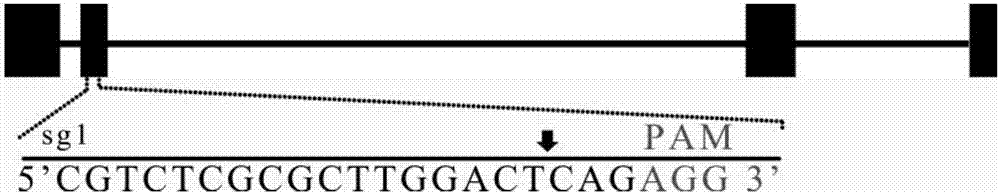
[0028] (1) sgRNA design
[0029] According to the genome sequence of porcine IGFBP3 gene (gene ID: 001005156), a sgRNA targeting porcine IGFBP3 gene was designed. The 20nt oligonucleotide sgRNA guide sequence is: IGFBP3-sgRNA1:5'-CGTCTCGCGCTTGGACTCAG-3', located in the second exon of the gene IGFBP3; add CACC to its 5' end to obtain the forward oligonucleotide sequence; according to The targeting sequence gets its corresponding DNA complementary strand, and AAAC is added to its 5' end to get the reverse oligonucleotide. Synthesize the above forward oligonucleotides and reverse oligonucleotides respectively, and synthesize the forward and reverse sgRNA oligonucleotide sequences: denature at 95°C for 5 minutes, and anneal at 72°C for 10 minutes; after annealing, sticky ends can be formed The specific oligonucleotide sequence is shown in Table 1.
[0030] Oligonucleotide sequence of table 1 sgRNA guide sequence
[0031]
Nucleotide sequence (5' to 3')
Sg1-F ...
Embodiment 2
[0037] (1) Acquisition of G418 resistance gene
[0038] Primers were designed according to the G418 resistance gene in the plasmid pEGFP-C1 vector, and a BsmBI restriction site was added to the 5' end of the primer oligonucleotide sequence, Kana-F: 5-GGCGTCTCCGGGATGTGCGCGGAACCCCTATTTG-3, Kana-R: 5-GGCGTCTCGCGCCAATTCTAAAGTATATGAGTAACC-3, Synthesize the upstream and downstream primer sequences respectively, and use the plasmid pEGFP-C1 as a template for PCR amplification. The PCR reaction system is 50 μL: pEGFP-C1 plasmid 1 μL, 2×PCRMix 25 μL, upstream and downstream primers 1 μL each, ddH 2 O 22 μL. The PCR reaction conditions were: 94°C for 5min, 94°C for 30s, 60°C for 45s, 72°C for 1min40s, a total of 33 cycles, followed by an extension at 72°C for 10min. The obtained DNA fragment was named Kana.
[0039] Table 2 Amplifies the primer sequence of Kana fragment
[0040]
Sequence (5' to 3')
Kana-F
GGCGTCTCCGGGATGTGCGCGGAACCCCTATTTG
Kana-R
G...
Embodiment 3
[0044] (1) Culture of porcine fibroblasts
[0045] For methods of isolation and culture of pig fetal fibroblasts, please refer to literature: Xinjige; Cheng Wenmin; Pan Weirong; Qing Yubo; Zha Xingqin; Fu Guowen; Wei Hongjiang; Zeng Yangzhi. Research on culture methods of pig fetal fibroblasts and ear skin fibroblasts, Heilongjiang Animal Husbandry and Veterinary Medicine, 2013(9): 175-179.
[0046] (2) Plasmid transfection and positive cell selection
[0047] The recombinant plasmid pX330-G418-IGFBP3-Sg1 was transfected into pig fetal fibroblasts by liposome transfection to obtain recombinant cells. For specific steps of transfection, refer to the operation manual of Liposome 3000 (Invitrogen, catalog number: 11668019) for transfection.
[0048] The transfected cells were screened with 1000ng / mL G418 for 10 days, and the screened cells were cultured and frozen.
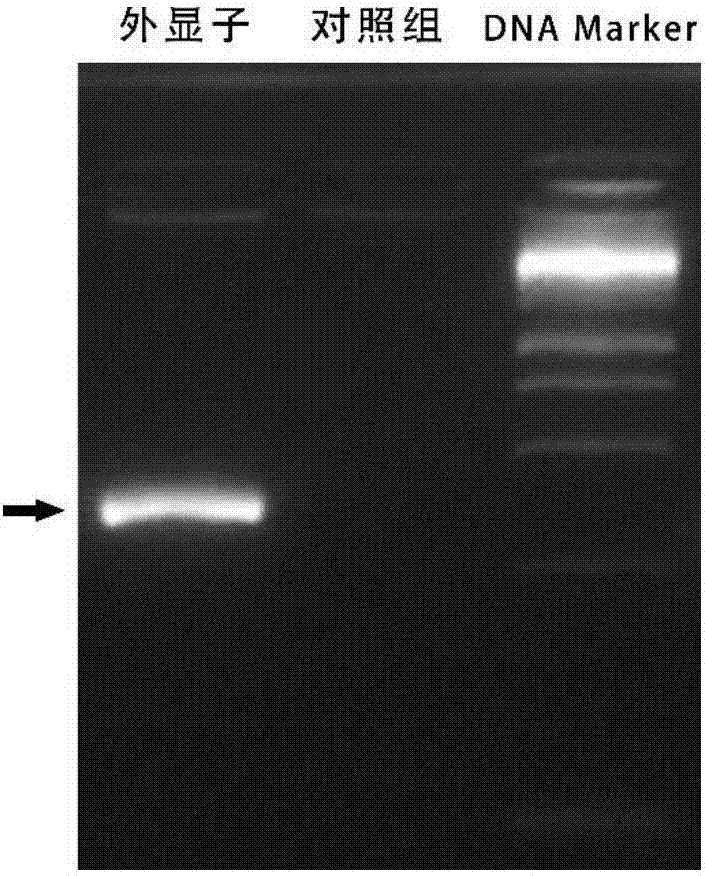
[0049] (3) Identification of gene knockout effect
[0050] Design primers for identification according to the ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com