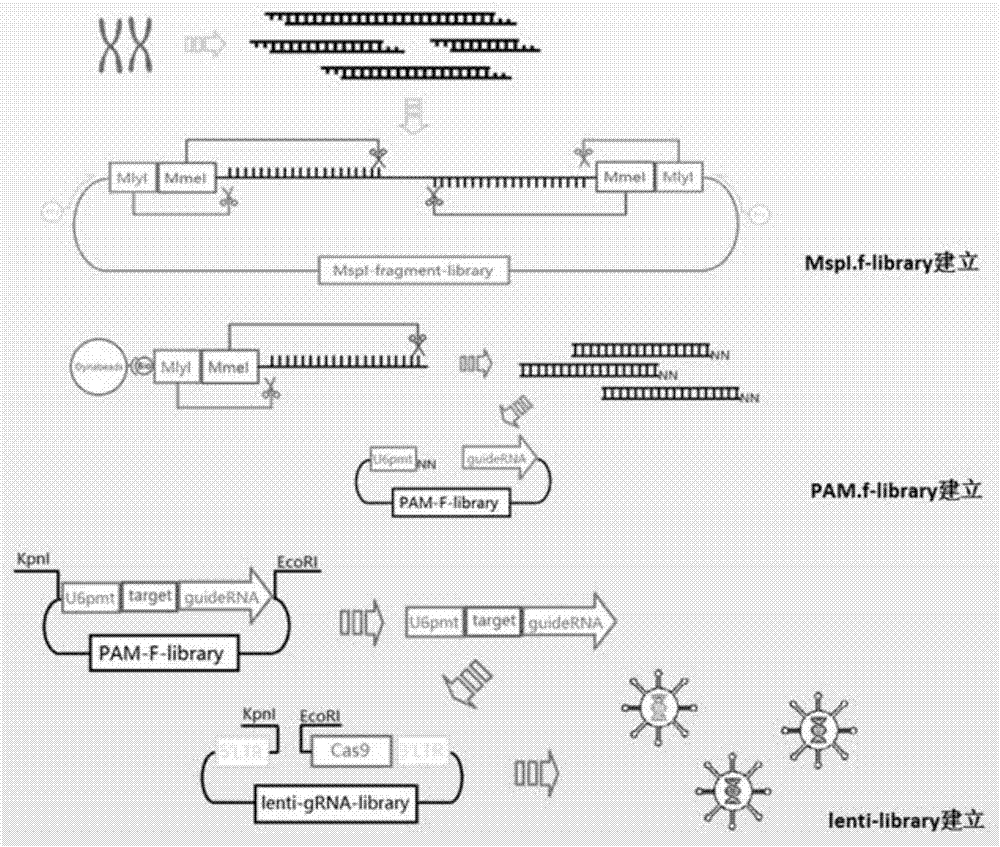
Method used for construction of CRISPR/Cas9 genome knockout library with enzymatically digested genome
A genome and library technology, applied in the field of genome knockout library construction, can solve the problems of time-consuming and labor-intensive, low coverage of manual design, imperfect genetic information of species, etc., to reduce production costs, improve coverage, and maintain stability Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0047] In this embodiment, the method for constructing a CRISPR / Cas9 genome knockout library by enzymatically cutting the genome comprises the following steps:
[0048] 1. Screening of cells stably expressing green fluorescent protein
[0049] The commercial pEGFP-C1 plasmid was transfected into the PK-15 cell line by liposome transfection, and G418 was used to screen, and the clones stably expressing green fluorescent protein were picked and continued to be cultured, and finally the stable expression green fluorescent protein was obtained. The PK-15 cell line was named PKpG-Pi.
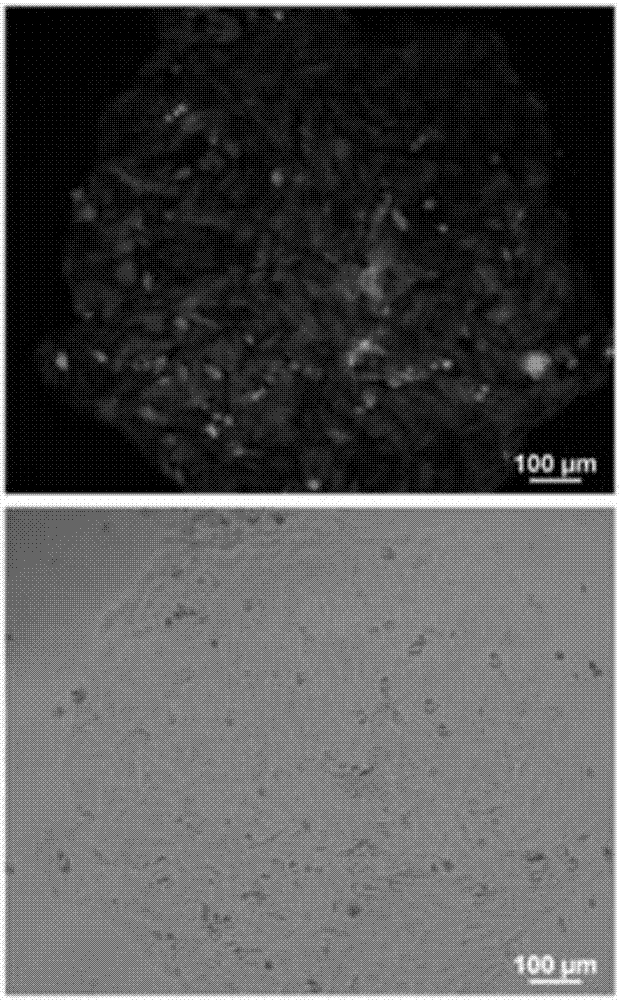
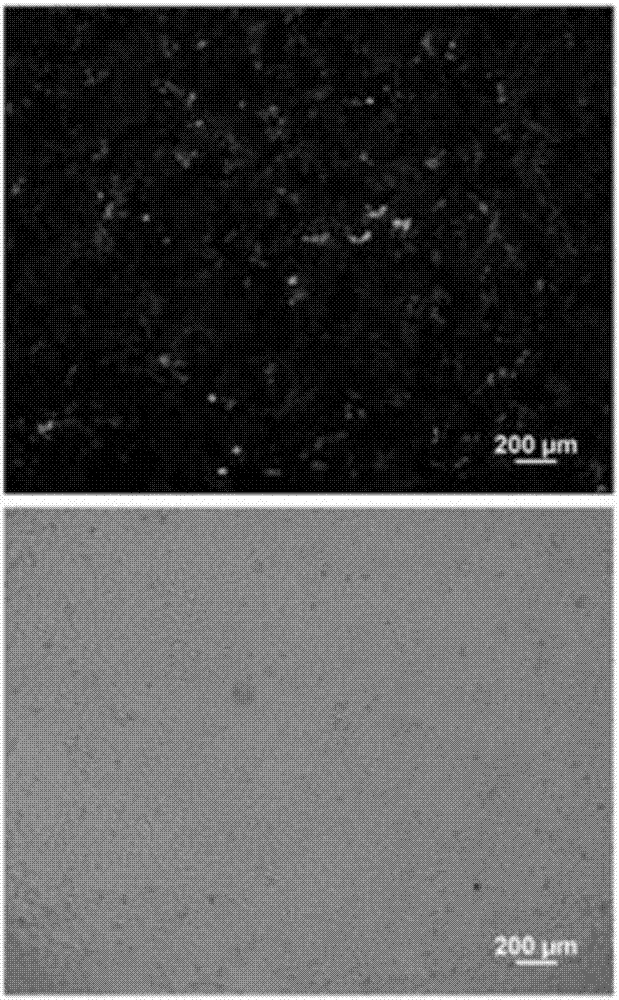
[0050] A clone of PK-15 cells expressing green fluorescent protein observed under a fluorescent microscope and an optical microscope is as follows: figure 2 as shown, figure 2 The upper part is the photo under the fluorescence microscope, and the lower part is the photo under the light microscope. A PKpG-Pi cell line stably expressing green fluorescent protein observed under a fluorescent micros...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com