A low-complexity prediction method for naturally disordered proteins
A low-complexity, prediction method technology, applied in the field of bioinformatics, can solve the problems of high computational complexity and low accuracy, achieve high computing speed and robustness, reduce the number of features and computational complexity.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
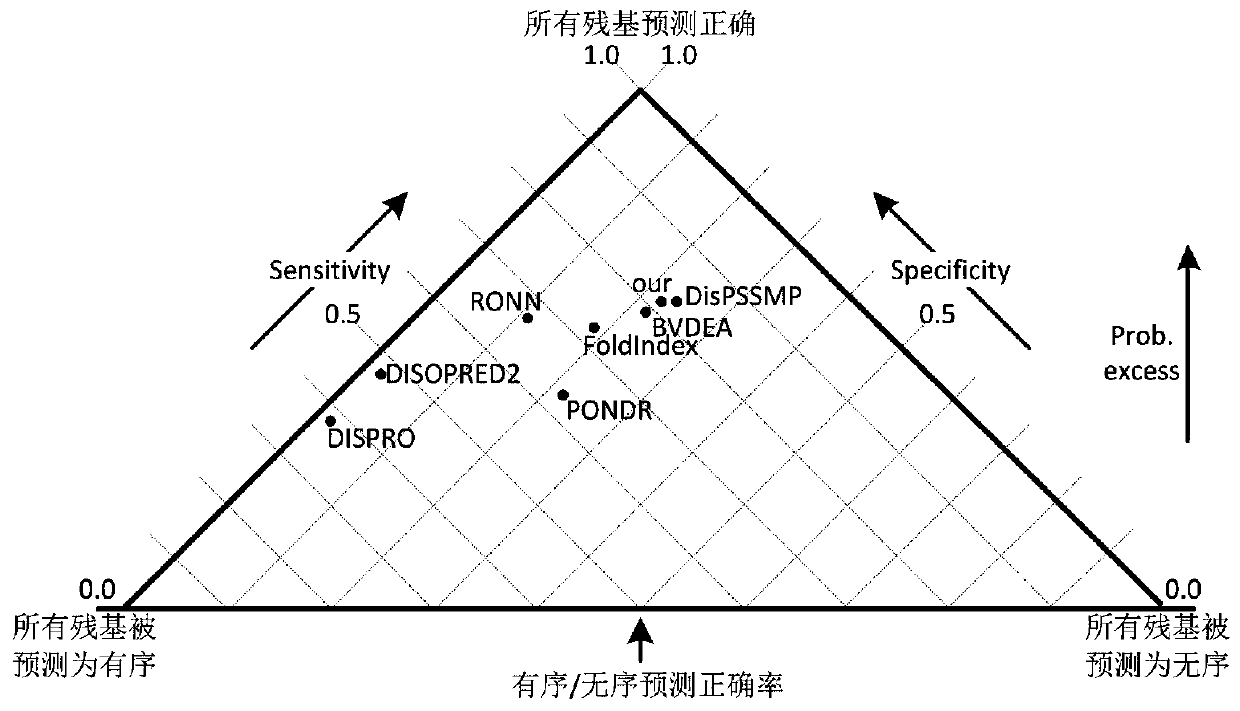
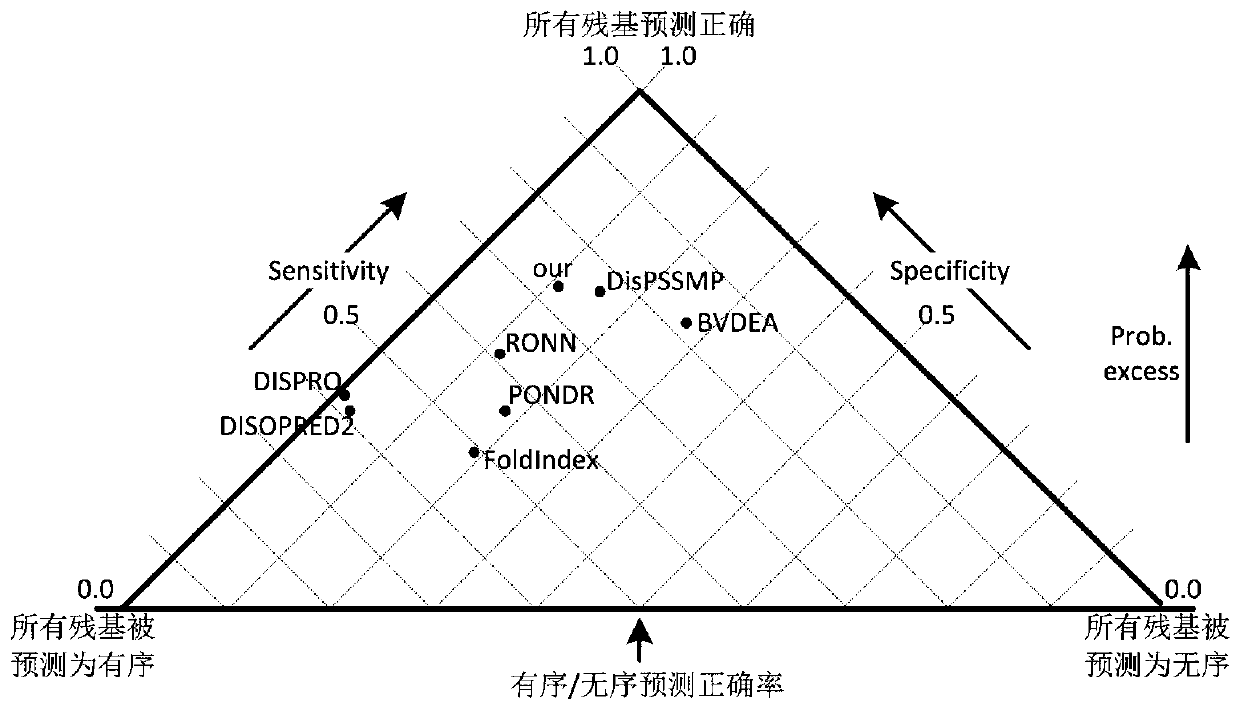
Image
Examples
Embodiment 1
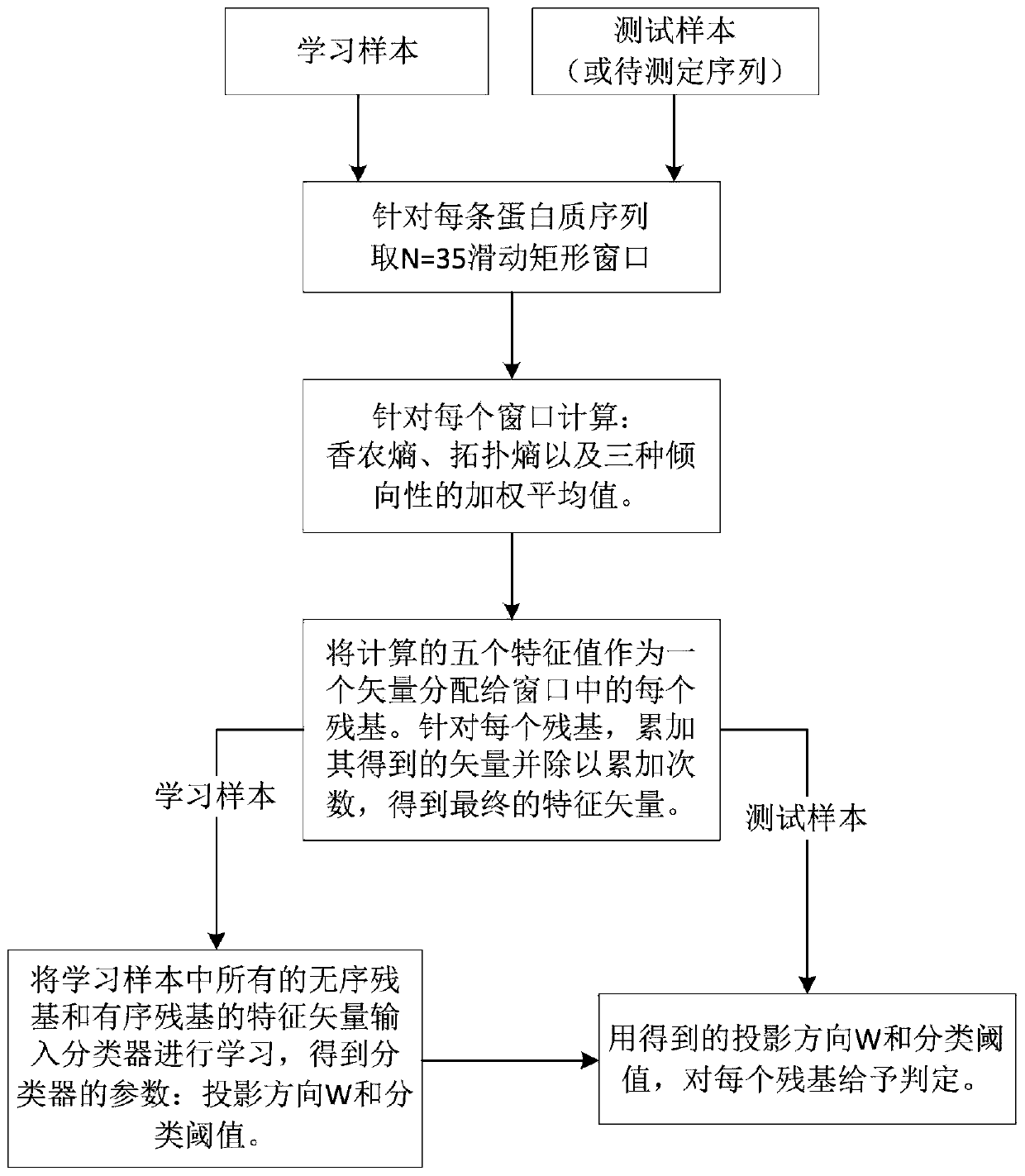
[0036] The specific steps of the method for predicting natural disordered proteins provided by the invention are as follows:
[0037] For a protein sequence w in an undetermined disordered region (taking a protein sequence labeled 1g4m in the R80 data set as an example), the specific steps for prediction using the disordered protein prediction scheme provided by the present invention are as follows:
[0038] Step 1: The length of the sequence is 393, and the sequence is intercepted with a sliding window of N=35. The values of five features are calculated for each window interval.
[0039] Sequence w = MGDKGTRVFKKASPNGKLTVYLGKRDFVDHIDLVEPV
[0040] For the first window with a length of N, according to the formula (1)(3)(4), calculate the values of the five features of the sequence fragments intercepted by the window, and assign these five values to each of the fragments Residues; after that, sliding window, calculate the value of the five features of the sequence fr...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com