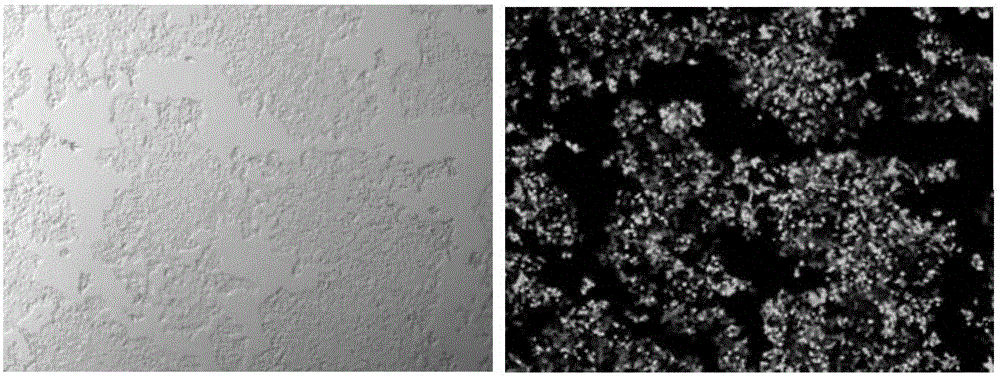
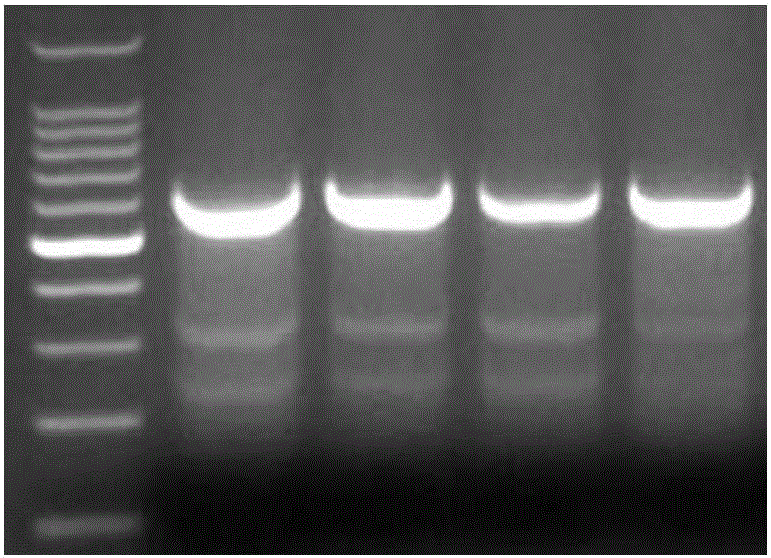
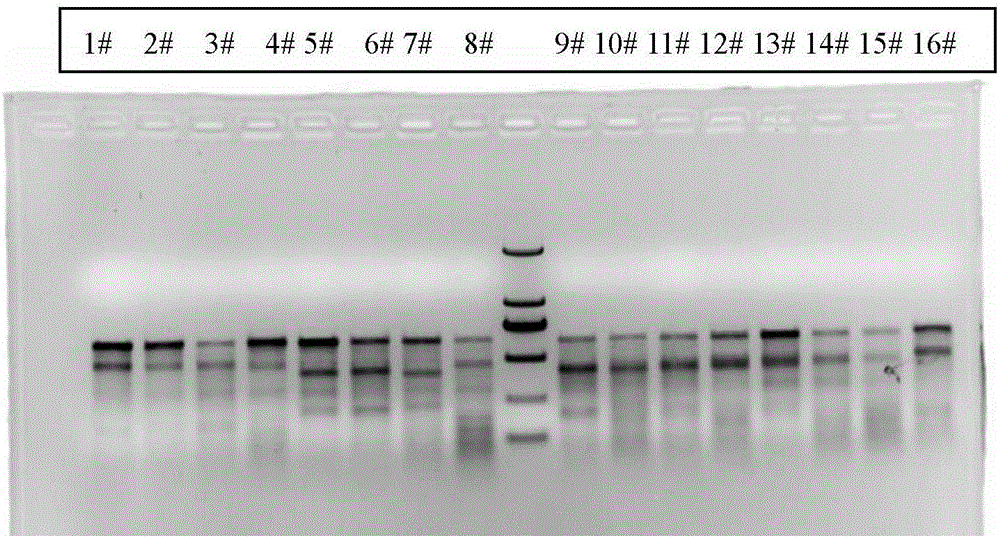
Sgrna (ribonucleic acid) sequence for editing ccr5 gene by crispr (clustered regularly interspaced short palindromic repeats) technology and application thereof
A DNA sequence, ccr5-sgrna2 technology, applied in DNA/RNA fragments, genetic engineering, recombinant DNA technology, etc., can solve problems such as non-homologous end joining insertion or deletion errors, and achieve high cutting efficiency and low off-target probability Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0020] The present invention will now be described more fully with reference to the accompanying drawings, in which some, but not all embodiments of the invention are depicted. Indeed, these inventions may be embodied in many different forms and should not be construed as limited to the embodiments shown herein as modifications and other embodiments are intended to be included within the scope of the appended claims.
[0021] 1. sgRNA
[0022] In general, a sgRNA is any polynucleotide sequence that is sufficiently complementary to a target polynucleotide sequence to hybridize to the target sequence and direct specific binding of the CRISPR complex to the target sequence.
[0023] In some embodiments, the sgRNA of the present invention can be further modified so that the modified sgRNA has the same sequence as listed above CCR5-sgRNA1: AGGGCAACTAAATACATTCT, CCR5-sgRNA2: TGCCAAAAAATCAATGTGAA, CCR5-sgRNA3: AGTGGGACTTTGGAAATACA, and CCR5-sgRNA4: ATGCACAGGGTGGAACAAGA has about 100...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com