Quantitative detection of gii-type norovirus in fruits and vegetables by one-step microdroplet digital PCR
A micro-droplet digital and quantitative detection technology, which is applied in the determination/inspection of microorganisms, biochemical equipment and methods, etc., to achieve the effects of good primer specificity, good stability, and sensitive detection methods
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0055] Example 1 RT-ddPCR amplification annealing temperature optimization
[0056] Synthesize the primer probe according to the upstream primer and probe sequence of Norovirus GII type in Loisy and the downstream primer of Norovirus GII type in Kageyama (see Note 1, 2):
[0057] Upstream primer QNIF2: 5′-ATGTTCAGRTGGATGAGRTTCTCWGA-3′;
[0058] Downstream primer COG2R: 5′-TCGACGCCATTCTTCATTCACA-3′;
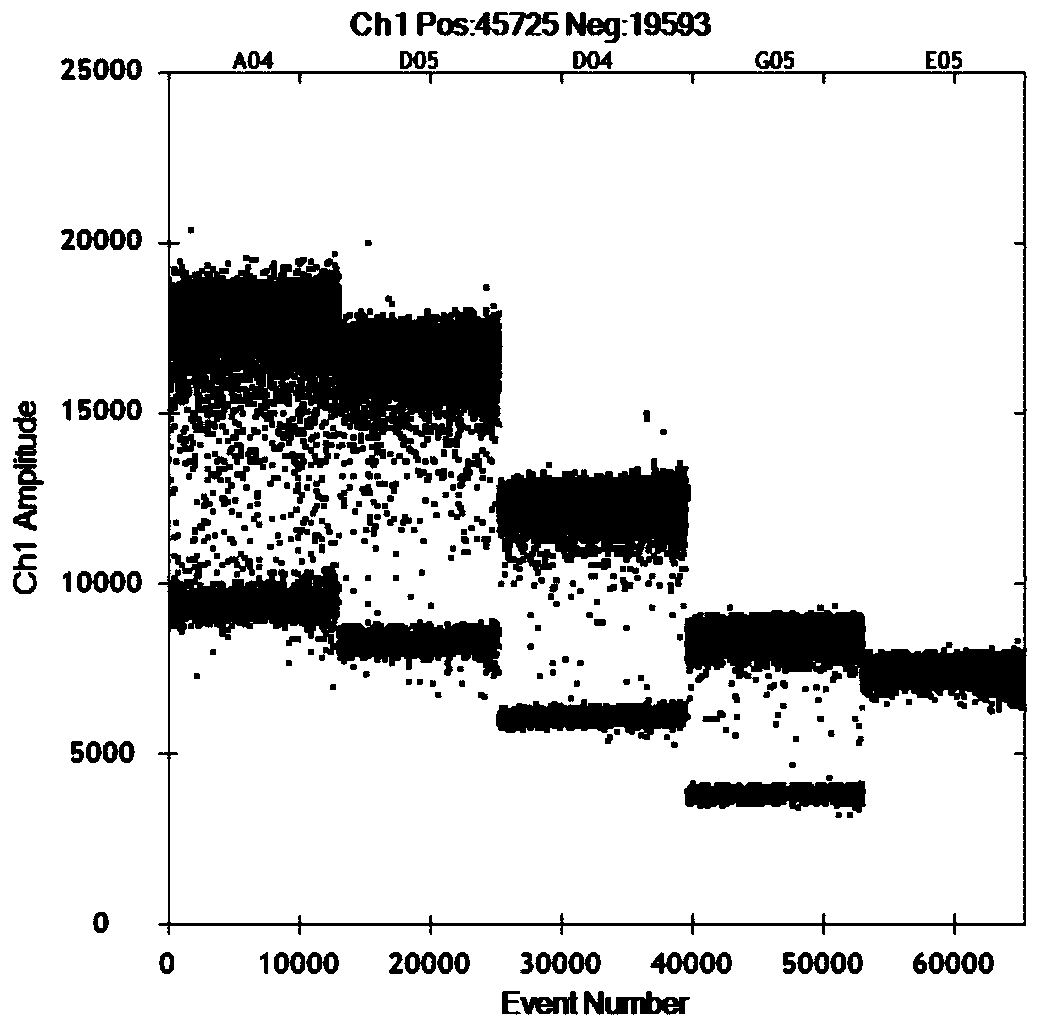
[0059] Probe QNIFs: 5′-AGCACGTGGGAGGGCGATCG-3′, the 5′ end of the probe is labeled with the fluorescent reporter group FAM, and the 3′ end is labeled with the fluorescent quencher group BHQ1. Using the same viral nucleic acid as a template, select 8 temperatures from 50 to 60°C (specifically 50°C, 51°C, 52°C, 54°C, 56°C, 57°C, 58°C, 60°C) according to the gradient temperature setting of the instrument Perform experimental optimization. The reaction conditions were: 5 μL of super mix, 2 μL of RT-Enzyme Mix, 1 μL of DTT in Bio-Rad’s One-step RT-ddPCR Advanced Kit for Probes kit, ...
Embodiment 2
[0062] Example 2 RT-ddPCR amplification primer concentration optimization
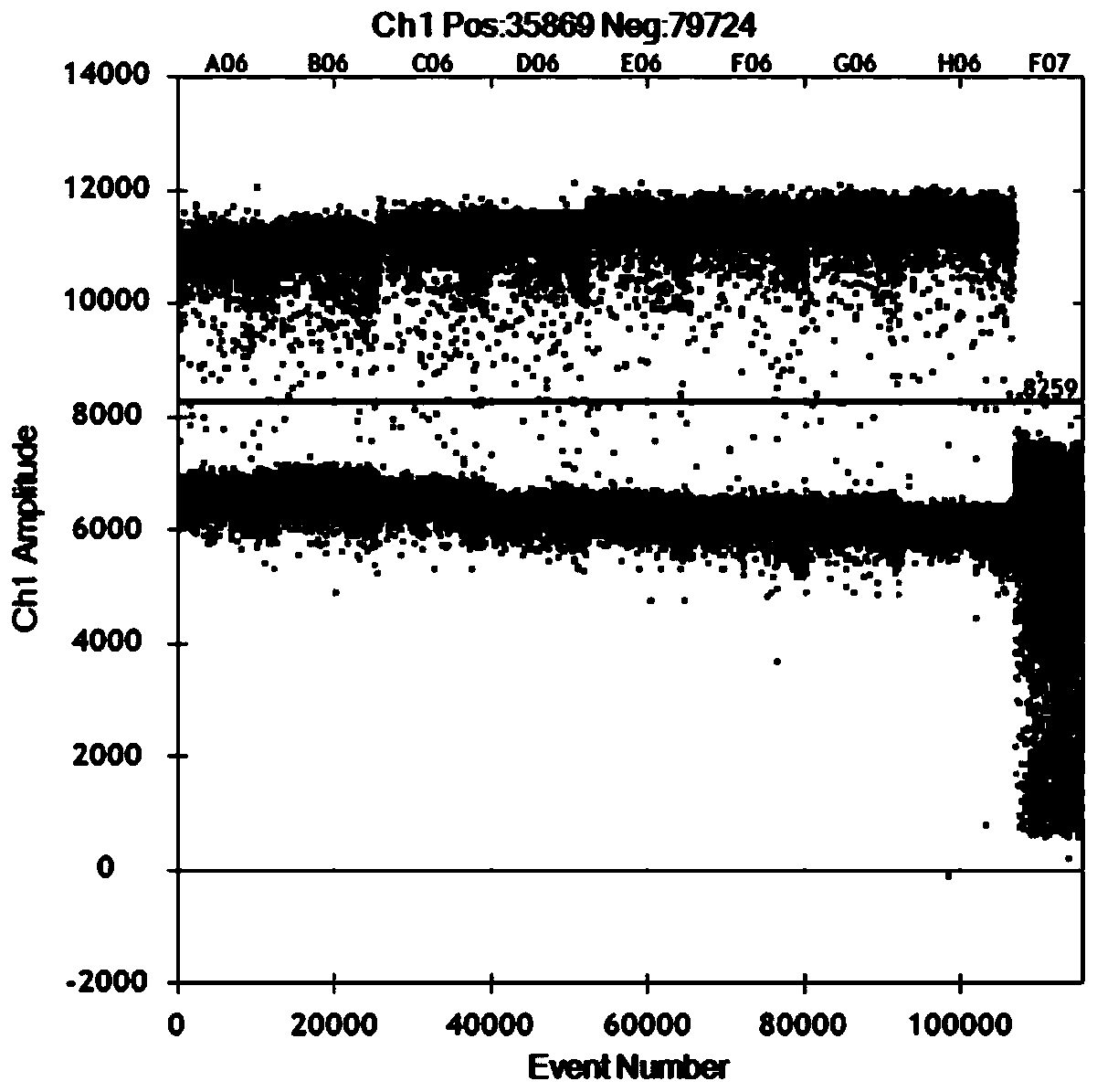
[0063] The same primer probe sequence as in Example 1 was used for the experiment, and the same viral nucleic acid was used as a template, and the primer concentrations of 0.3 μM, 0.6 μM, 0.9 μM, and 1.2 μM were selected for investigation, and the remaining reaction conditions were: Bio-Rad One-step In the RT-ddPCR Advanced Kit for Probes kit, 5 μL of super mix, 2 μL of RT-Enzyme Mix, 1 μL of DTT, the final concentration of 0.35 μmol / L probe concentration, 2 μL of template addition, supplemented with RNase Free water to 20 μL. Reverse transcription was performed at 42°C for 60 minutes, 95°C for 10 minutes to inactivate reverse transcriptase, 95°C for 30 seconds, 54°C for 1 minute, 45 cycles, 98°C for 10 minutes, and the ramp rate was 1°C / s. The results show that the reaction detection result reaches the maximum value in the 0.9 μM primer concentration, so the primer concentration of 0.9 μM ( figure 2...
Embodiment 3
[0064] Embodiment 3RT-ddPCR amplification probe concentration optimization
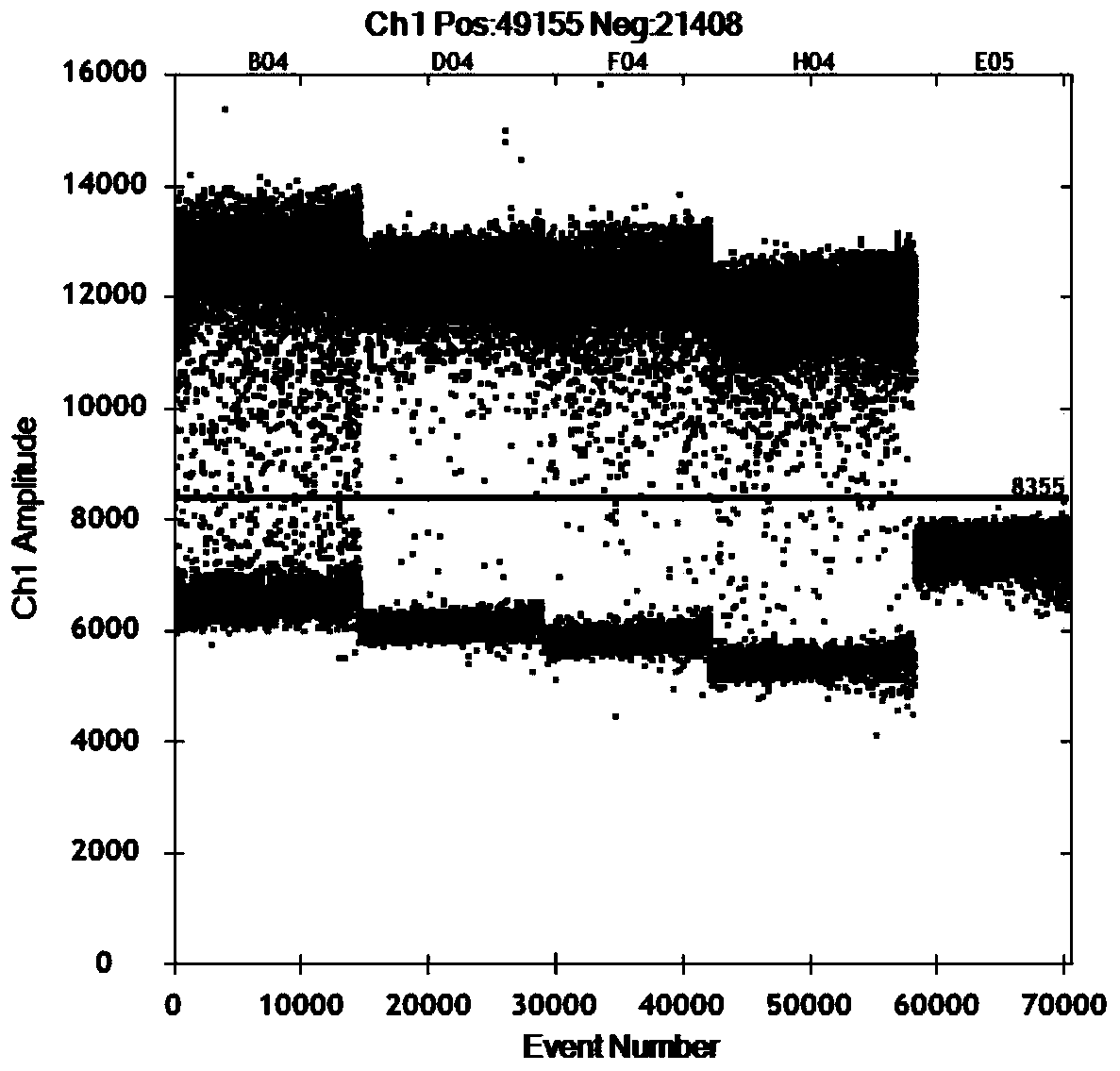
[0065] The same primer probe sequence as in Example 1 was used for the experiment, and the same viral nucleic acid was used as a template, and the probe concentrations of 150nM, 250nM, 350nM, and 450nM were respectively selected for investigation, and the remaining reaction conditions were: Bio-Rad One-step RT-ddPCR In the Advanced Kit for Probes kit, 5 μL of super mix, 2 μL of RT-Enzyme Mix, 1 μL of DTT, the final concentration of 0.9 μM primer concentration, 2 μL of template addition, supplemented with RNase Free water to 20 μL. Reverse transcription was performed at 42°C for 60 minutes, 95°C for 10 minutes to inactivate reverse transcriptase, 95°C for 30 seconds, 54°C for 1 minute, 45 cycles, 98°C for 10 minutes, and the ramp rate was 1°C / s. The result shows that the reaction detection result reaches the maximum in the 350nM probe concentration, so the probe concentration of 350nM ( image 3 ).
PUM
| Property | Measurement | Unit |
|---|---|---|
| PCR efficiency | aaaaa | aaaaa |
| PCR efficiency | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com