Yeast Engineering Strain Transformed by Chromosomal Fusion
A technology for yeast engineering and engineering strains, applied in genetic engineering, stable introduction of foreign DNA into chromosomes, microorganism-based methods, etc., and can solve problems such as limiting the application of heterologous gene expression
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
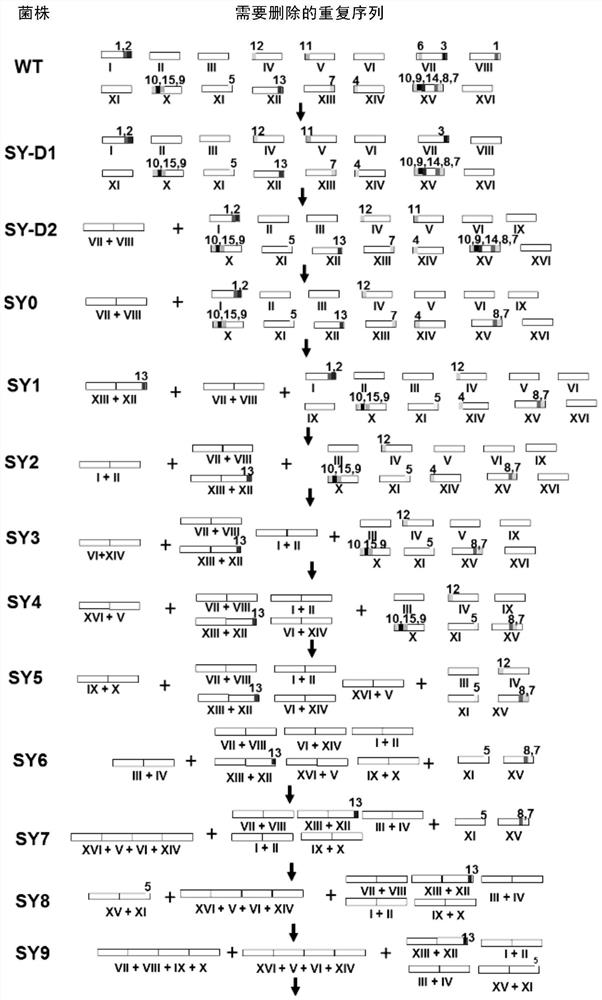
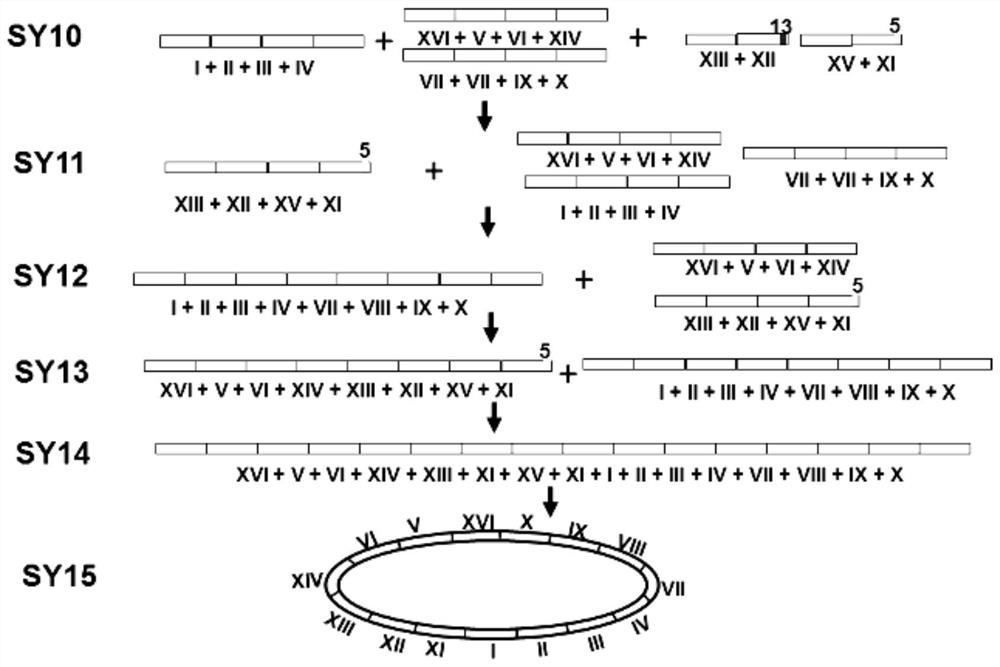
[0212] Example 1 , strain construction
[0213] The starting strain of a series of chromosomally fused engineering strains constructed by the present invention is the derivative strain BY4742 of Saccharomyces cerevisiae wild-type strain S288C, and its genotype is MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 (Brachmann, C.B., etc. (1998); Yeast, 14, 115- 132).
[0214] There are 13 major repeat sequences in the proximal telomeric region of the 16 natural chromosomes of strain BY4742 (Table 1), of which repeat sequences 1, 7, 9, and 10 are 3 copies, and the remaining 11 repeat sequences are 2 copies. The length of these repeated sequences is 2.7-26.2kb, and they tend to become hotspots of chromosome homologous recombination, thereby affecting the stability of the entire genome, and also interfere with the site-directed homologous recombination of chromosome fusion. In order to eliminate the main repeat sequence near the telomere, the inventors deleted a total of 19 repeat sequences, incl...
Embodiment 2
[0232] Example 2 , strain performance
[0233] Strain growth
[0234] A series of chromosomal fusion engineered strains of Saccharomyces cerevisiae constructed by the invention have good growth. The one-by-one fusion of linear natural chromosomes did not affect the growth of the strain. Figure 5 The growth curve of the strain SY11-SY13 ( Figure 5 , A) and SY14 ( Figure 5 , B) The growth was almost identical to that of the wild-type starting strain BY4742. The circularization of the chromosome made the growth of the strain SY15 slightly slower than that of the wild-type strain BY4742 ( Figure 5 , B).
Embodiment 3
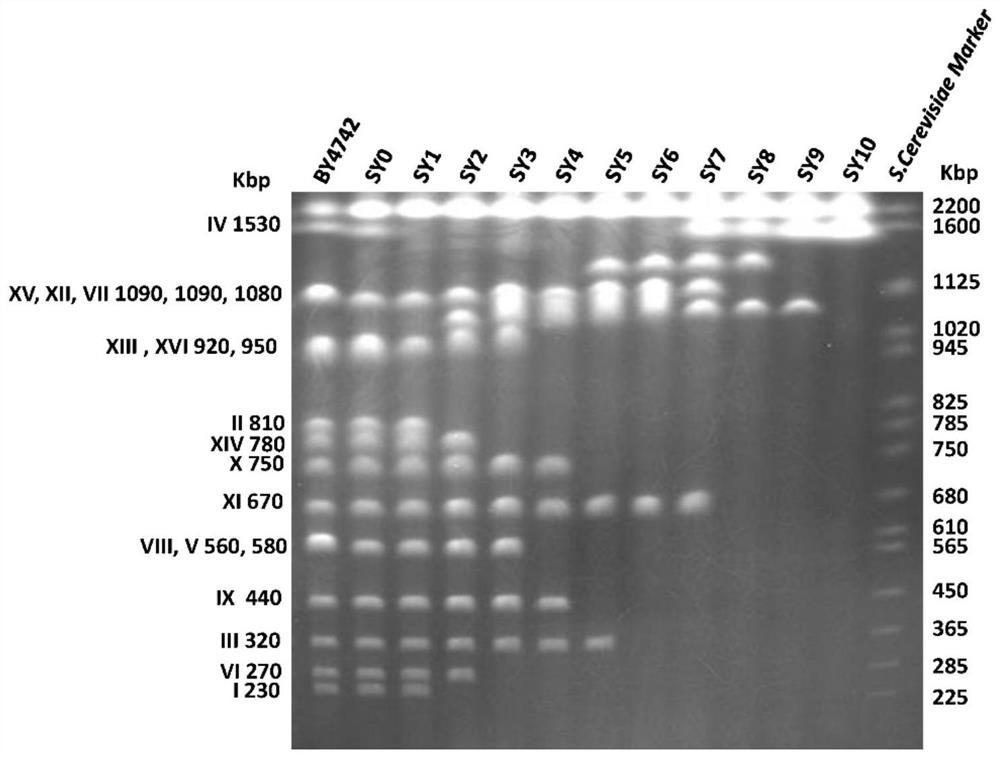
[0235] Example 3 , genome stability
[0236] A series of chromosomal fusion engineered strains of Saccharomyces cerevisiae constructed by the invention have genetically stable genomes. Image 6 Shown are the enzyme digestion verification pictures of the genomes of strains SY14 and SY15 after 0 generation and continuous growth of 100 generations. The chromosome of strain SY14 can theoretically cut out 23 bands with restriction endonuclease FseI, which are 1569, 1446, 1125, 1050, 892, 891, 778, 587, 548, 414, 412, 381, 363, 318, 258 , 210, 141, 133, 82, 78, 27, 25.7, and 0.9 kb. The chromosome of the strain SY15 is derived from the circularization of the chromosome of SY14, so comparing the FseI restriction map of SY15 with that of SY14, two differential bands of 1125 and 82 kb will be reduced. These two bands were merged into one in SY15, that is, a new 1193kb differential band was added.
[0237] The de novo sequencing whole genome sequence of strain SY14 is shown in SEQ ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com