Method of detecting breast cancer early diagnosis related genes based on nucleic mass spectrometry
A technology of technical detection and nucleic acid mass spectrometry, which is applied in the field of detecting genes related to early diagnosis of breast cancer based on nucleic acid mass spectrometry technology, which can solve the problems of low detection amount, high background, artificial artifacts of DNA chain secondary structure, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
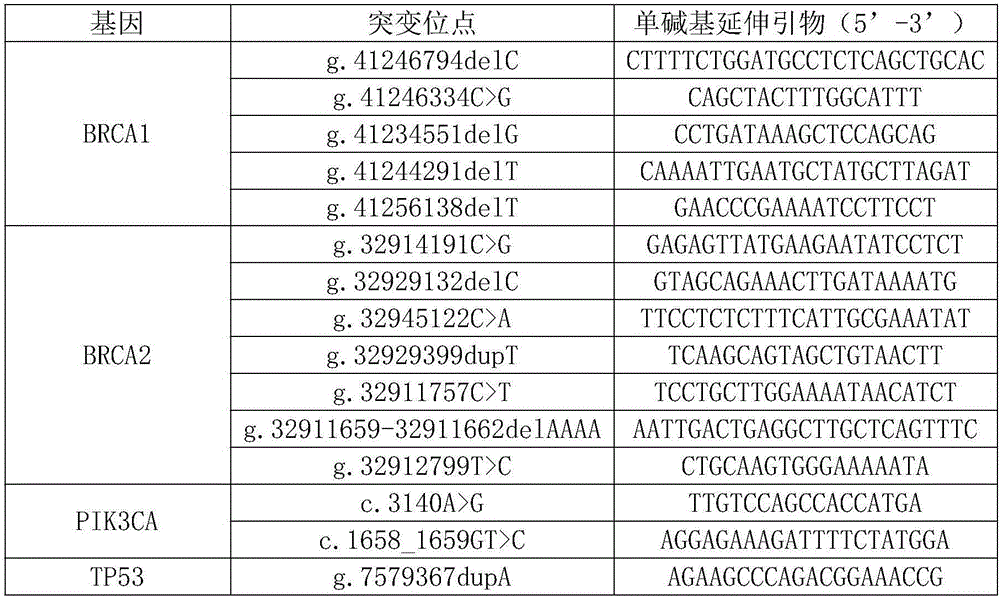
[0135] Example 1: Detection of breast cancer susceptibility-related SNP sites and gene mutation sites by the technology of the present invention
[0136] 1. Extract DNA from peripheral blood samples to obtain DNA extraction solution;
[0137] 1. Take 200μl peripheral blood sample and put it into a 1.5ml centrifuge tube. (If the initial volume of whole blood is less than 200μl, use buffer BB to make up to 200μl. If the initial volume is between 200μl-300μl, then increase the reagent volume proportionally in subsequent operations. If the initial volume is between 300μl-1ml lysing of red blood cells is required first)
[0138] 2. Add 200 μl of binding solution CB, shake it upside down vigorously immediately, and mix well, then add 20 μl proteinase K (20 mg / ml) solution, shake it upside down and mix well, place at 70°C for 10 minutes, the solution should become clear.
[0139] 3. Add 100 μl of isopropanol, shake vigorously upside down, and mix well. At this time, flocculent prec...
Embodiment 2
[0196] Embodiment 2: Detection of breast cancer cell lines by the technical scheme of the present invention
[0197] 1. Research object
[0198] Six liver cancer cell lines (all available from ATCC) with known mutation sites MCF7, MDA-MB-361, CAMA-1, MDA-MB-468, UACC-3199, and MDA-MB-231 were selected for this test To analyze the feasibility of the method, set up normal human genome DNA (gDNA) (extracted from the oral mucosa tissue of normal people in Wuhan General Hospital of Guangzhou Military Region) as a negative control, and water (H 2 0) as a blank control.
[0199] 2. Experimental steps
[0200] (1) extracting breast cancer cell line DNA to obtain a DNA extract;
[0201] (2) As shown in Table 5, 12 pairs of amplification primers of P1 group, 6 pairs of amplification primers of P2 group, 9 pairs of amplification primers of P3 group and 6 pairs of amplification primers of P4 group are mixed respectively, and then The mixed 4 sets of specific amplification primers were...
Embodiment 3
[0219] Example 3: From the peripheral blood of 26 pathologically diagnosed breast cancer patients, detect a group of SNPs and mutation sites of genes related to early diagnosis of breast cancer
[0220] 1. Experimental steps
[0221] A group of SNPs and mutation sites of genes related to early diagnosis of breast cancer were detected in the peripheral blood of 26 patients with pathologically diagnosed breast cancer obtained from Wuhan General Hospital of Guangzhou Military Region. The specific steps are as follows:
[0222] (1) Obtain the subject's cfDNA sample using the Biotech Whole Blood Genomic DNA Rapid Extraction Kit
[0223] (a) Take 200 μl of collected peripheral blood and put it into a 1.5ml centrifuge tube. Add 200 μl of binding solution CB, shake it upside down vigorously immediately, and mix well, then add 20 μl proteinase K (20 mg / ml) solution, shake it upside down and mix well, place at 70°C for 10 minutes, the solution should become clear (but the color is blac...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com