Modification of transcriptional repressor binding site in NF-YC4 promoter for increased protein content and resistance to stress
A protein content, NF-YC4 technology, applied to cells modified by introducing foreign genetic material, applications, chemical instruments and methods, etc., can solve problems such as unexplained functions
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
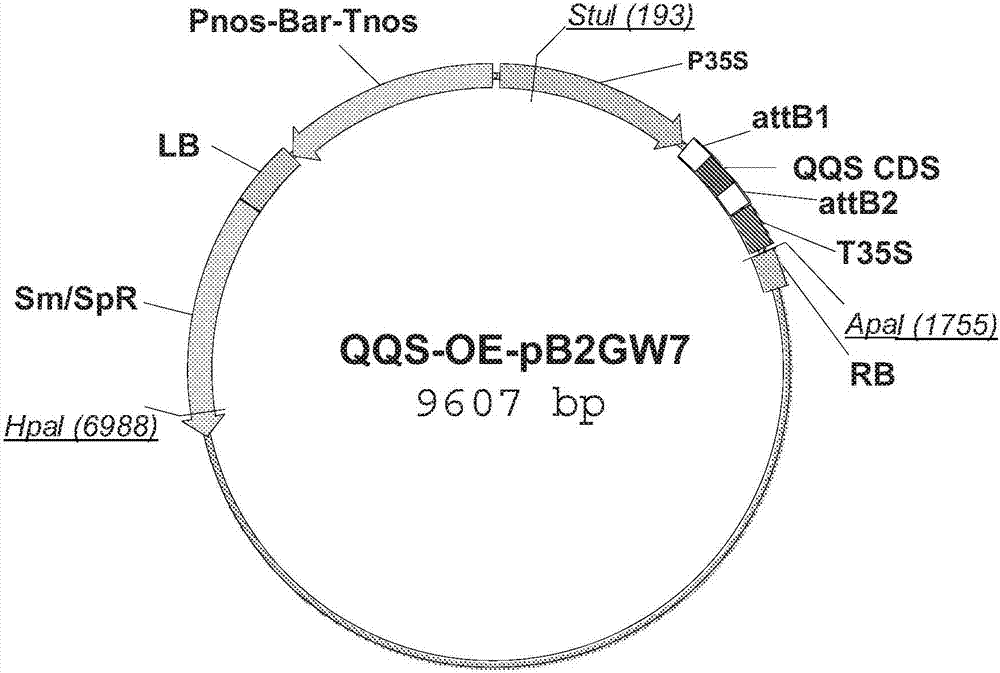
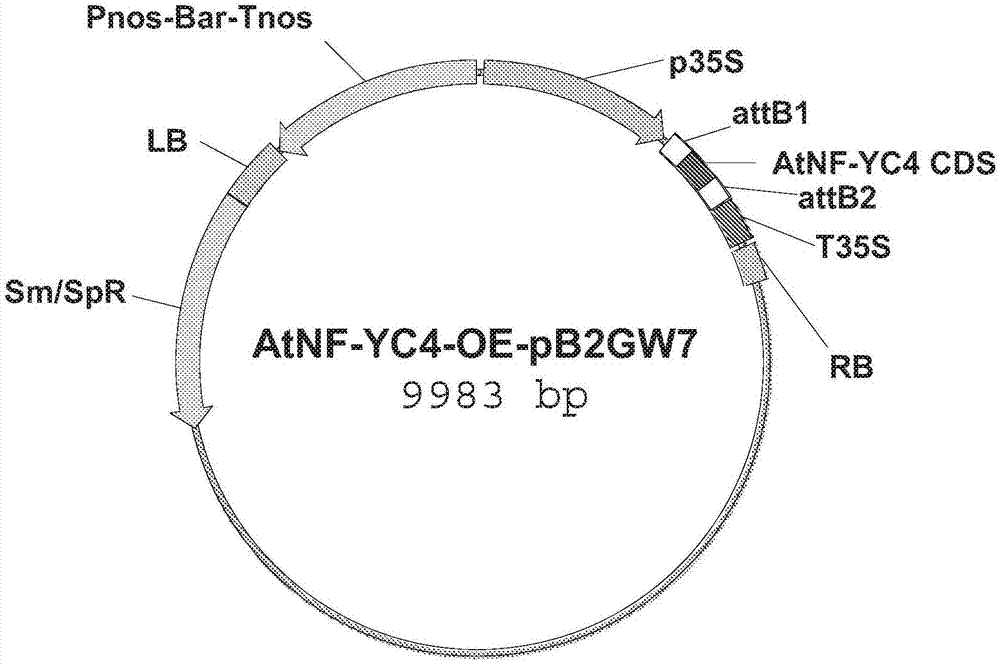
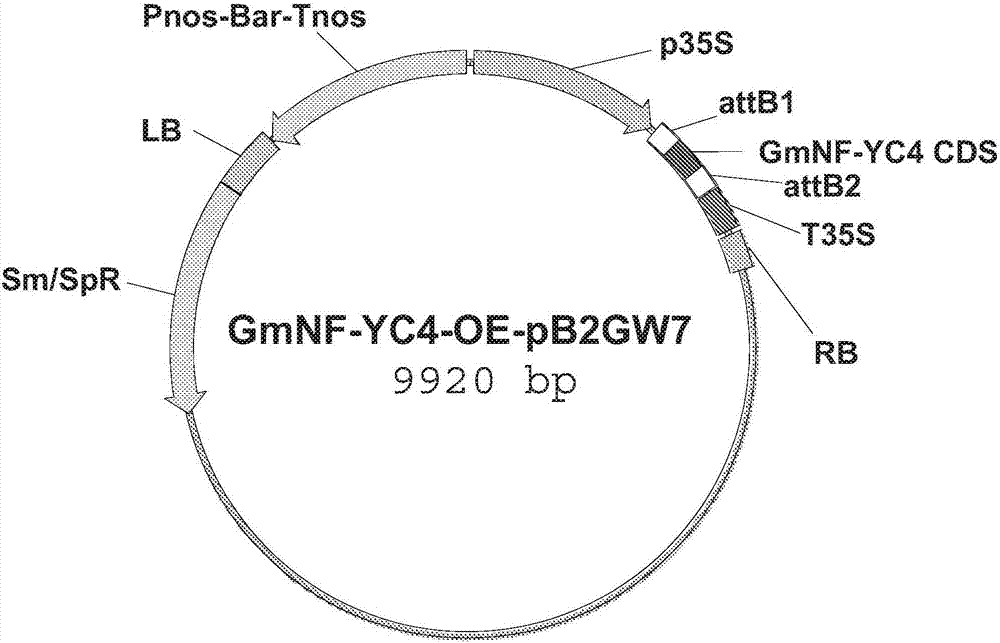
[0132] This example describes the preparation and transformation of 35S::QQS and 35S::AtNF-YC4 fusion constructs.
[0133] The 35S::QQS and 35S::AtNF-YC4 fusion constructs were made by cloning the amplified full-length coding sequence into the binary vector pB2GW7, as described above (Li et al. (2015), supra). Briefly, the QQS coding domain sequence (CDS) or the NF-YC4 CDS from Arabidopsis thaliana (AtNF-YC4) was inserted into the vector pB2GW7 (see Figures 1a and 1b for vector maps), which contains the Bar gene ( glufosinate acetyltransferase gene) under the control of Pnos (nos promoter) and Tnos (nos terminator) of nopaline synthase from tobacco (Nicotiana tabacum). The CDS on this vector is controlled by p35S (Cauliflower Mosaic Virus (CaMV) 35S promoter) and terminated by T35S (CaMV 35S). Only the region between LB and RB is introduced into plants to generate transgenic plants.
[0134] The constructs were introduced into Agrobacterium tumefaciens strain GV3101 for tran...
Embodiment 2
[0141] This example describes the analysis of plant composition.
[0142] Determination of leaf composition (starch staining and quantification and protein), in Arabidopsis, in seedling shoots 20 days after planting (DAP) in a growth chamber, and in rice, once in plants from 30 DAP Measured on the fallen leaves (next to the flag leaves) of the branches. 3 (for starch) or 5 (for protein) plants per replicate, 3 replicates from each independent T2 line (Arabidopsis) and T3 line (rice) were analyzed. Leaves were harvested at the end of the light phase. Seeds of Arabidopsis, rice and soybean were harvested from individual plants. For leaf and seed protein quantification, plant material was roasted at 71°C. Dried leaf / seed tissue (0.07 g) was used for each assay. Measure total protein content. Total nitrogen content was determined with LECO CHN-2000 ((LECO, St. Joseph, MI) and converted into protein content (Li et al. (2015), supra).
[0143] The whole aerial part of Arabidop...
Embodiment 3
[0146] This example describes a yeast two-hybrid assay.
[0147] The Matchmaker System 3 (Clontech, Mountain View, CA) was used to identify QQS-interacting proteins using an Arabidopsis (type Columbia) cDNA library constructed from 3-day-old etiolated seedlings (Arabidopsis.org / servlets / TairObject?type=library&id =23). For the interactive yeast two-hybrid assay, QQS and AtNF-YC4 were cloned into pGBKT7 (bait vector) and pGADT7 (trap vector), respectively.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com