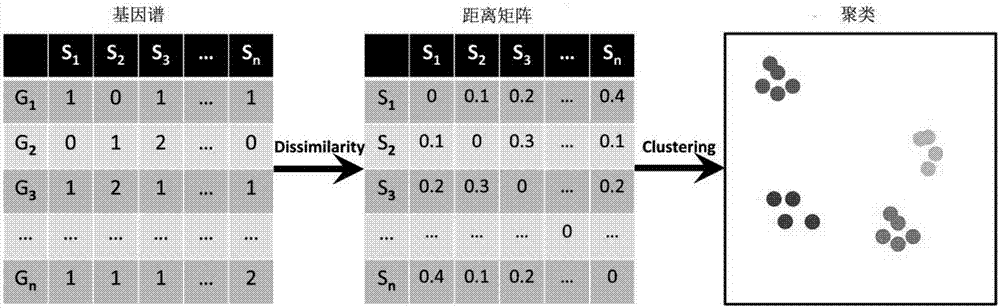
High-similarity microorganism identifying and classifying method
A classification method and microbial technology, applied in special data processing applications, instruments, electrical digital data processing, etc., can solve the problems of inability to accurately identify and classify highly similar microorganisms, and achieve high accuracy and good application prospects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
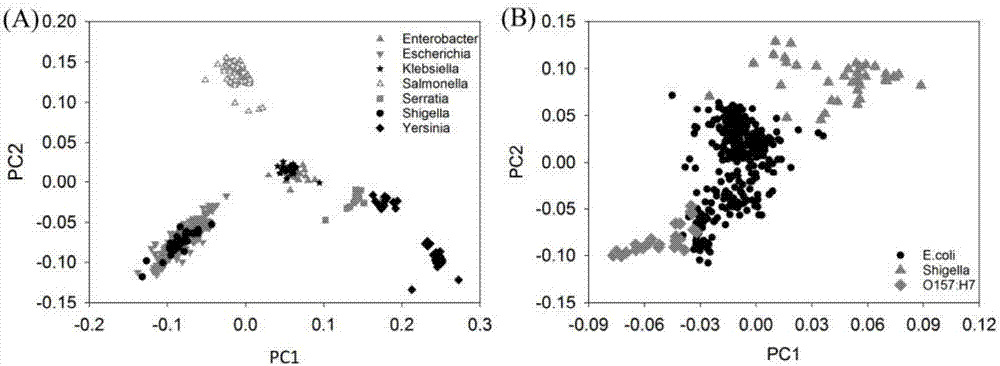
[0028] Identification and classification of embodiment 1 Enterobacteriaceae
[0029] 1) Strain collection
[0030] In this case, a total of 916 sequenced Enterobacter genomes were collected, including 14 strains of Enterobacter, 384 strains of Escherichia, 45 strains of Klebsiella, 314 strains of Salmonella, and 14 strains of Serratia. There were 42 strains of Shigella and 103 strains of Yersinia pestis.
[0031] 2) Strain identification and classification
[0032] When all genera were mixed together for identification and classification, most genera could be classified better. However, it is difficult to distinguish between Shigella and Escherichia, and it is also difficult to distinguish between Enterobacter and Klebsiella, but the trend is obvious. However, Y. pestis and Y. pseudotuberculosis in Yersinia pestis were clearly distinguished from other Yersinia pestis species. These phenomena are basically consistent with the conclusions observed in clinical practice. Furt...
Embodiment 2
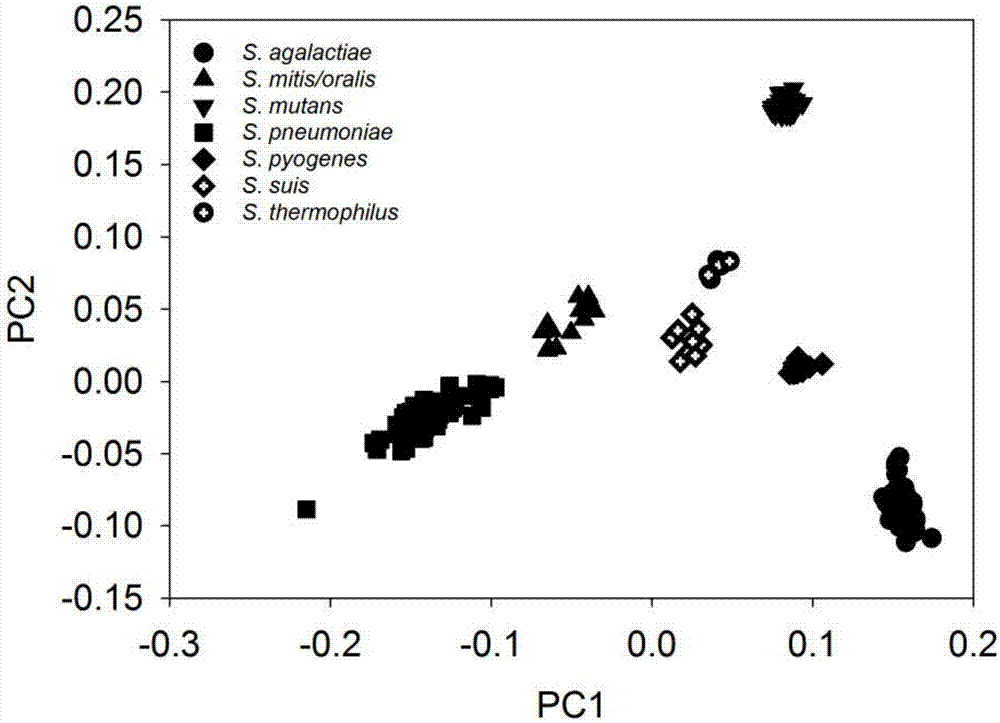
[0033] Identification and classification of embodiment 2 streptococci
[0034] 1) Strain collection
[0035] A total of 283 streptococcus strains were collected in this case, including 83 strains of S.agalactiae, 7 strains of S.mitis, 8 strains of S.oralis, 38 strains of S.mutans, 119 strains of S.pneumoniae, 12 strains of S.pyogenes, and 10 strains of S. .suis, and 6 strains of S.thermophilus.
[0036] 2) Strain identification and classification
[0037] Cluster analysis of the 283 Streptococcus strains showed that all species could be well distinguished, including S.oralis and S.mitis. S.oralis and S.mitis are highly similar in microbial taxonomy, and were once considered to be the same species unit. Based on the non-similarity clustering of gene content, the two types of streptococci have obvious distinctions, which can be considered to belong to different microbial species.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com