Residue interaction network comparison algorithm SI-MAGNA introducing sequence information
A technology for introducing sequences and sequence information, applied in the field of residue interaction network alignment algorithm SI-MAGNA, which can solve problems such as unseen algorithms and unseen residue interaction network alignment algorithms, and achieves improved accuracy and reasonableness. The effect of stability, high EC, high edge correctness
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
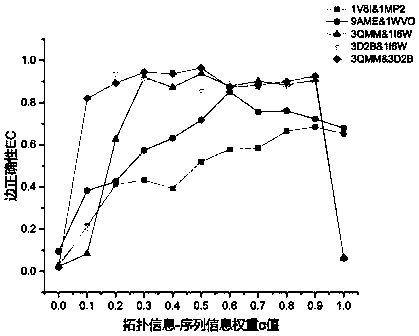
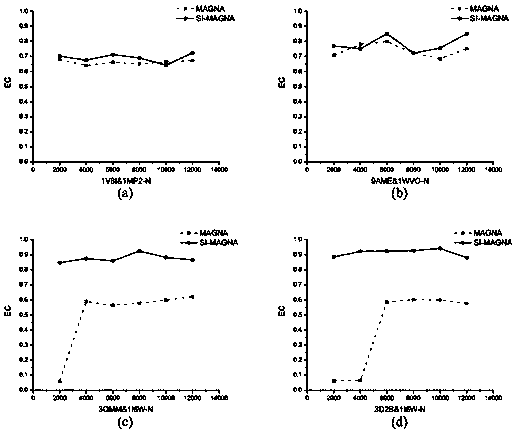
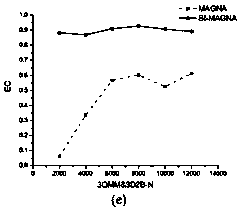
[0037] (1) In this paper, three groups of residue interaction network pairs are selected as the input of the algorithm, namely: (a) ADP-ribose pyrophosphatase from Thermus thermophilus (PDB number: 1V8I) and the structure from Mycobacterium tuberculosis Hydrolase (PDB number: 1MP2); (b) Type Ⅲ antifreeze protein isoform (PDB number: 9AME) from high-latitude temperate sea fish and sialic acid synthase (PDB number: 1WVO) from humans; ( c) Wild mesophilic lipase from Bacillus subtilis (PDB number: 1I6W) and its two thermophilic mutants (PDB number: 3D2B and 3QMM). The three-dimensional structure information and sequence information of different protein pairs were obtained from the RCSB PDB database (http: / / www.rcsb.org / pdb / home / home.do), and the BLAST algorithm was used for sequence alignment; and the residue-residue Based on the interaction of residues, the interaction network of residues is constructed, and then the network comparison is carried out.
[0038] (2) Use the SI-MA...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com