Methods and compositions for diagnosing or detecting lung cancers
一种组合物、肺癌的技术,应用在组合化学、生物化学设备和方法、生物信息学等方向,能够解决难以准确地预测、不适当调控等问题
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
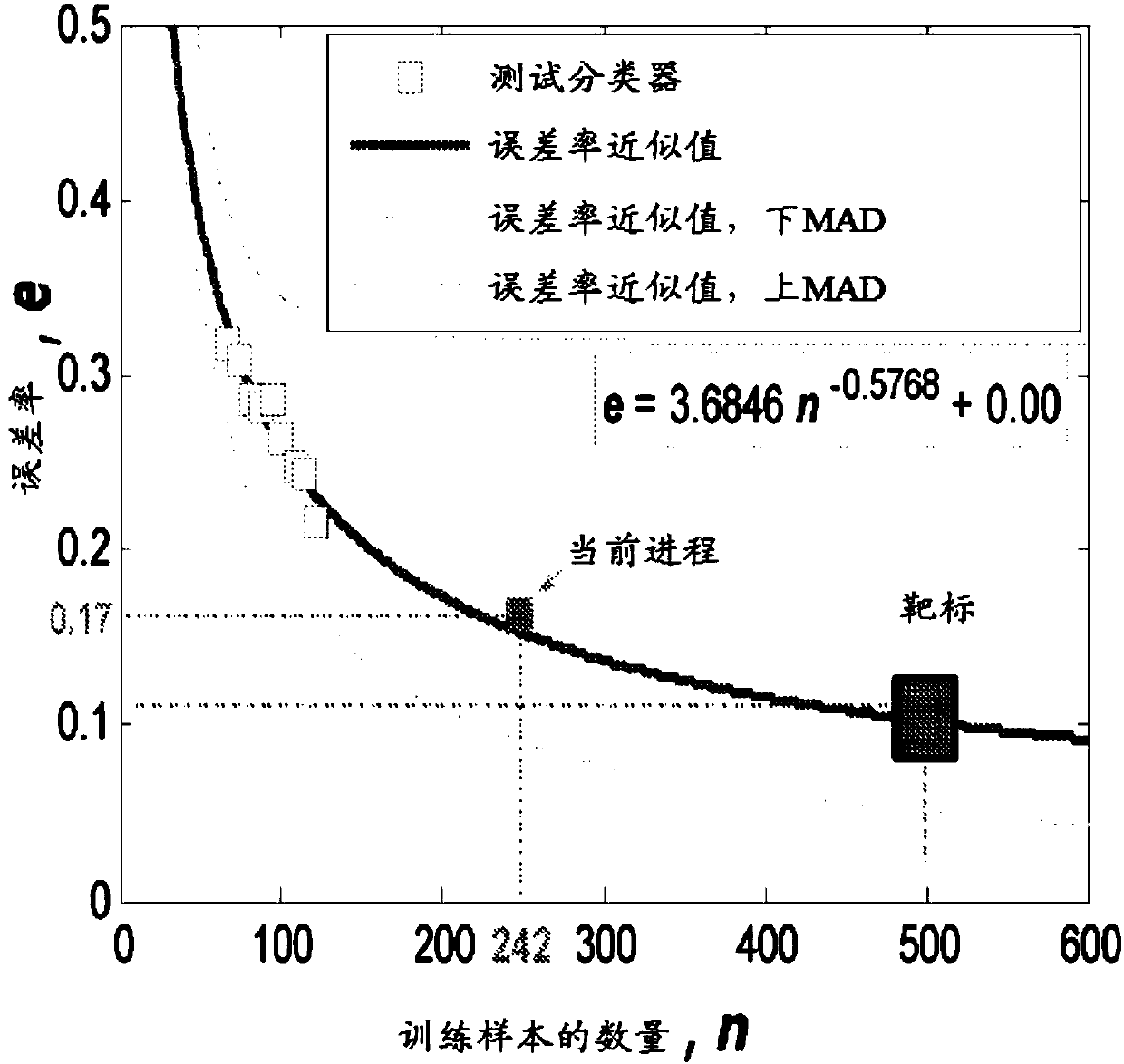
[0172] Example 1: Sample size calculation
[0173] The calculation is based on figure 1 PAXgene data as described in. We used data from the current PAXgene dataset of 23 cancer patients and 25 controls to design the sample size needed to achieve the desired 90% accuracy on the test set. We randomly selected training sets of different sizes with 24 to 44 samples, which is equivalent to 50% to 90% of all samples. The sample size is gradually increased in increments of 2 to allow one cancer sample and one control sample to be added at each step. For each given sample size, 50 resamples are performed.
[0174] Then a t-test was performed on each training set to identify the top 100 genes ranked by p value. The gene list is further reduced by removing any low expressors (expression not exceeding twice the average background level of all samples in the cancer and non-cancer groups).
[0175] The remaining 58 genes are then used to aggregate all samples, including those originally kept ...
Embodiment 2
[0177] Example 2-RNA purification and quality assessment
[0178] RNA purification for gene and miRNA array processing is performed using standard procedures such as the regular services of Genomics Core. Use of Qiagen that allows simultaneous purification of mRNA and miRNA TM The standard commercially available kit for the preparation of PAXgene RNA. The resulting RNA is used for mRNA or miRNA profile analysis.
[0179] RNA quality is measured using a biological analyzer. Use only RNA integrity value> 7.5 sample. A constant amount (100ng) of total RNA was amplified (aRNA) using an Illumina-approved RNA amplification kit (Epicenter). This program provides sufficient amplification material for multiple repeated gene and miRNA expression. If an alternative collection system will be used to collect smaller samples at a later date, RNA quantities as low as 10 ng can be used.
Embodiment 3
[0180] Example 3-Data preprocessing, array quality control, detection filtering
[0181] Array data is processed by Illumina’s Bead Studio, and the expression levels of signals and control probes are derived for analysis. In order to reduce experimental noise, the data was filtered by removing non-informative probes (probes that were not detected in more than 95% of all samples) and probes that did not vary by at least 1.2 times between any two samples. Then normalize the expression level quantile. These programs produce quantile-normalized data, and non-informative probe data is removed.
[0182] After each hybridization batch, we use all signal probes (> The expression level of 40,000) was calculated as the gene-wise global correlation as the median Spearman correlation on all microarrays, and the median absolute deviation of the global correlation was calculated. For each microarray, the median Spearman correlation is calculated relative to all other arrays, and arrays whose m...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com