A fusion protein and a method for homologous recombination using it
A technology of fusion protein and homologous recombination, applied in the field of fusion protein, can solve the problem of low efficiency of homologous recombination, achieve the effect of improving the efficiency of precise insertion, improving efficiency and reducing difficulty
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0061] Example 1. Design and construction of fusion protein spCas9-Avd
[0062] The coding sequences of CRISPR / spCas9 (hereinafter referred to as spCas9) and streptavidin Avidin are fused together through the linker sequence shown in SEQID NO: 1; spCas9 and Avidin share a start codon and a stop codon , for fusion expression, and the obtained fusion protein is referred to as spCas9-Avidin; the coding sequence of fusion protein spCas9-Avidin is shown in SEQ ID NO: 3, which has been verified by sequencing.
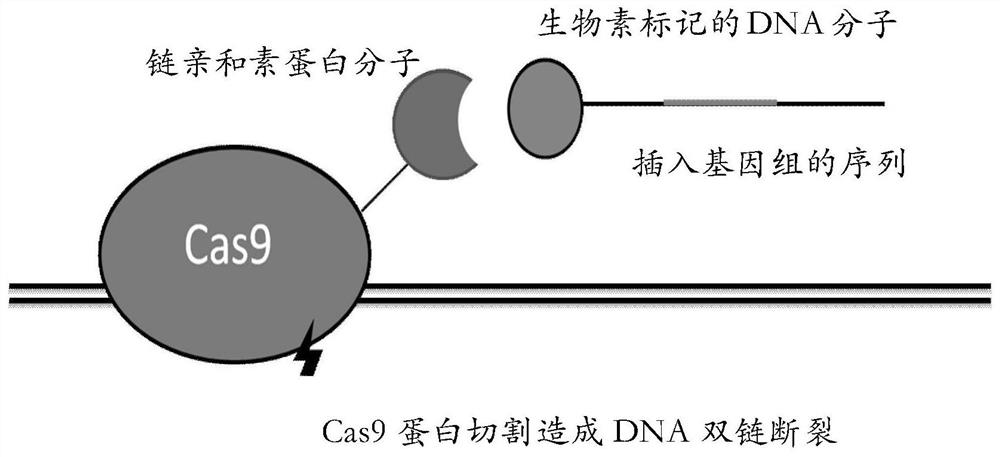
[0063] The inventor expects that the fusion protein can recruit the single-stranded DNA donor sequence with biotin modification at the 5' end to the double-stranded DNA break for homologous recombination repair. The schematic diagram of the principle is as follows. figure 1 shown. In order to verify this expectation, the inventors conducted the following experiments for verification.
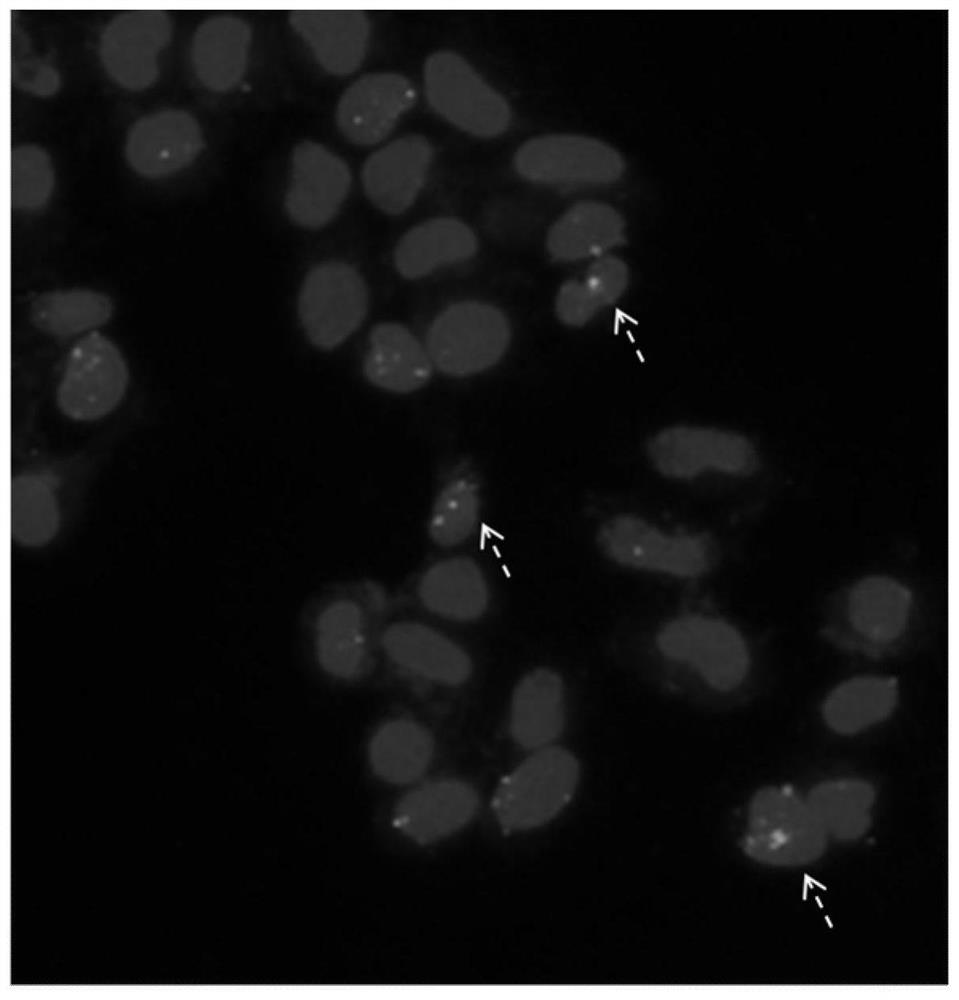
[0064] The inventors first tested whether the fusion protein can normally pull biotin-lab...
Embodiment 2
[0066] Example 2. The fusion protein spCas9-Avd significantly improves the homologous recombination efficiency at the cellular level
[0067] A. Experimental principle:
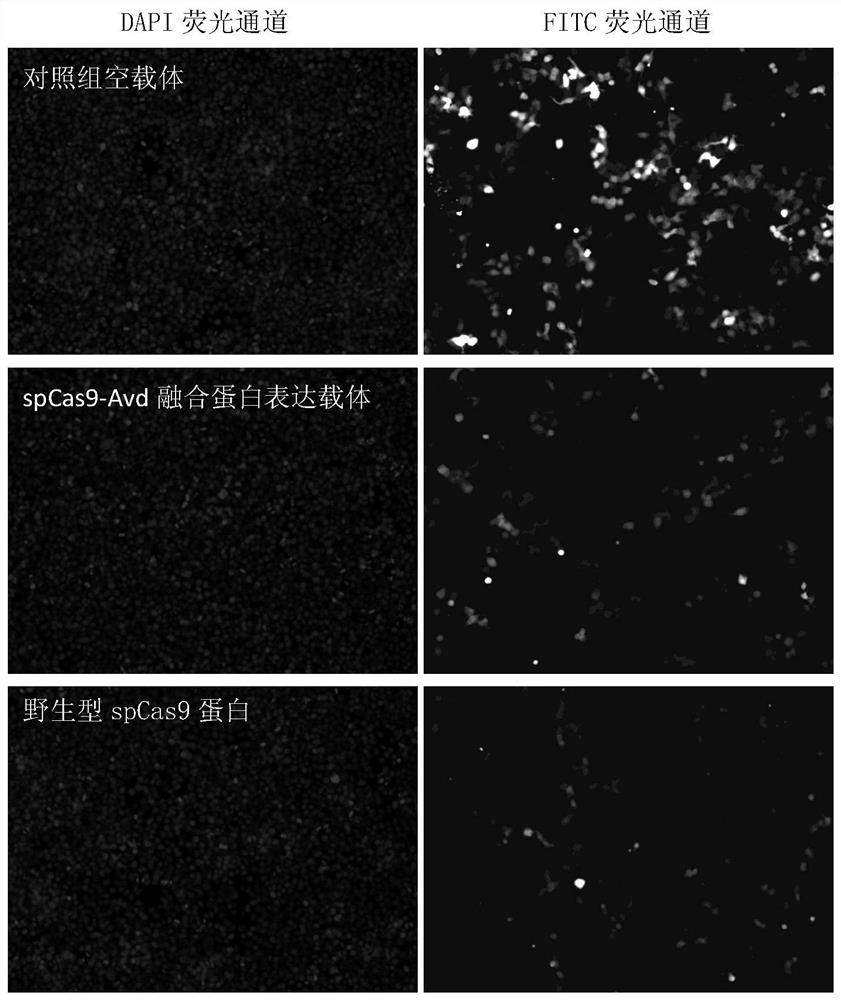
[0068] First, Sensor was integrated into the 293T cell genome; Sensor was the promoter followed by GFP with a stop codon TGA mutation. Normally, the cells do not fluoresce green. Then, the gRNA sequence is designed according to the target near the TGA. The donor single-stranded DNA contains left and right homology arms, and contains the sequence to repair the stop codon TGA into TGG; under the guidance of the gRNA, the CRISPR / Cas9 protein is located at the stop codon. Nearby cuts form double-stranded DNA breaks, and the donor single-stranded DNA has a certain probability to become a template for homologous recombination to complete the repair of TGA. Successful homologous recombination cells have green fluorescence, and the percentage of green fluorescent cells can be counted, which can then be used as an...
Embodiment 3
[0084] Example 3. The fusion protein spCas9-Avd significantly improves the production efficiency of knock-in mice
[0085] A. Experimental principle:
[0086] Objective: To select a sequence of the intron of Sirt7, the gene target of ICR mouse, as the target sequence, and to precisely knock a DNA sequence GGATCC (ie BamHI restriction site) into the genome.
[0087] Specific operations: The DNA sequence of the spCas9-Avidin fusion protein prepared in Example 1 was transcribed and capped by T7 enzyme; the designed corresponding gRNA plasmid was also transcribed into mRNA by T7; the donor single-stranded DNA was directly synthesized . The mixture of these three was microinjected into the fertilized mouse egg and transplanted into the uterus of the female mouse; about 12 days later, the developing mouse embryo was taken out, the genome was extracted after lysis, and the partial sequence of the target gene was passed through. PCR was performed for amplification; it was identifi...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com