Cpf1-based DNA in-vitro splicing method
A technology of motif and reaction system, applied in the field of DNA splicing in vitro based on Cpf1
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
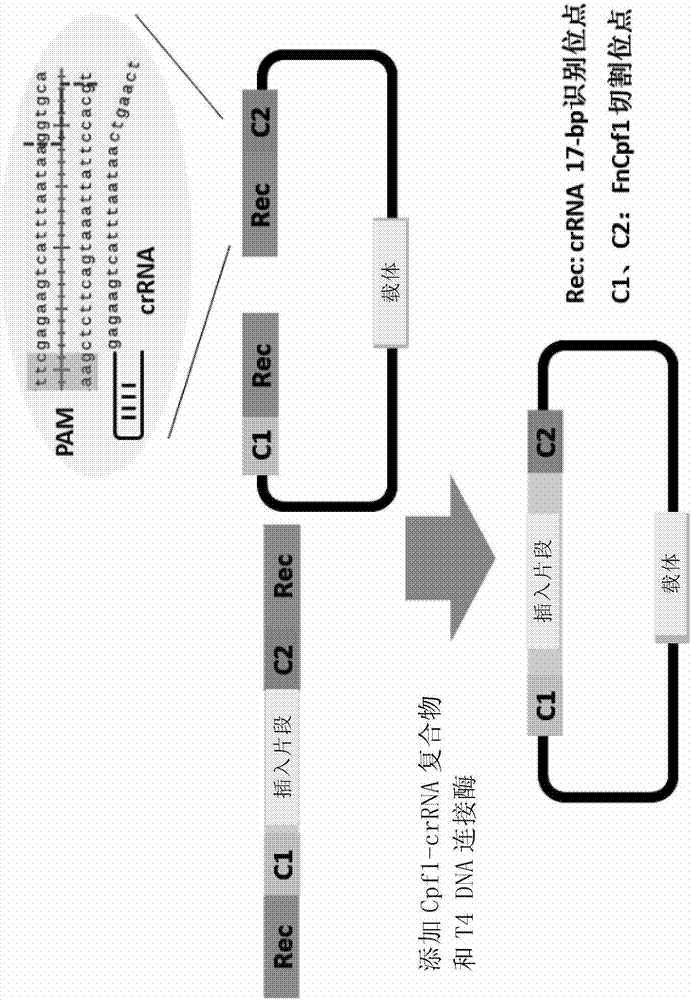
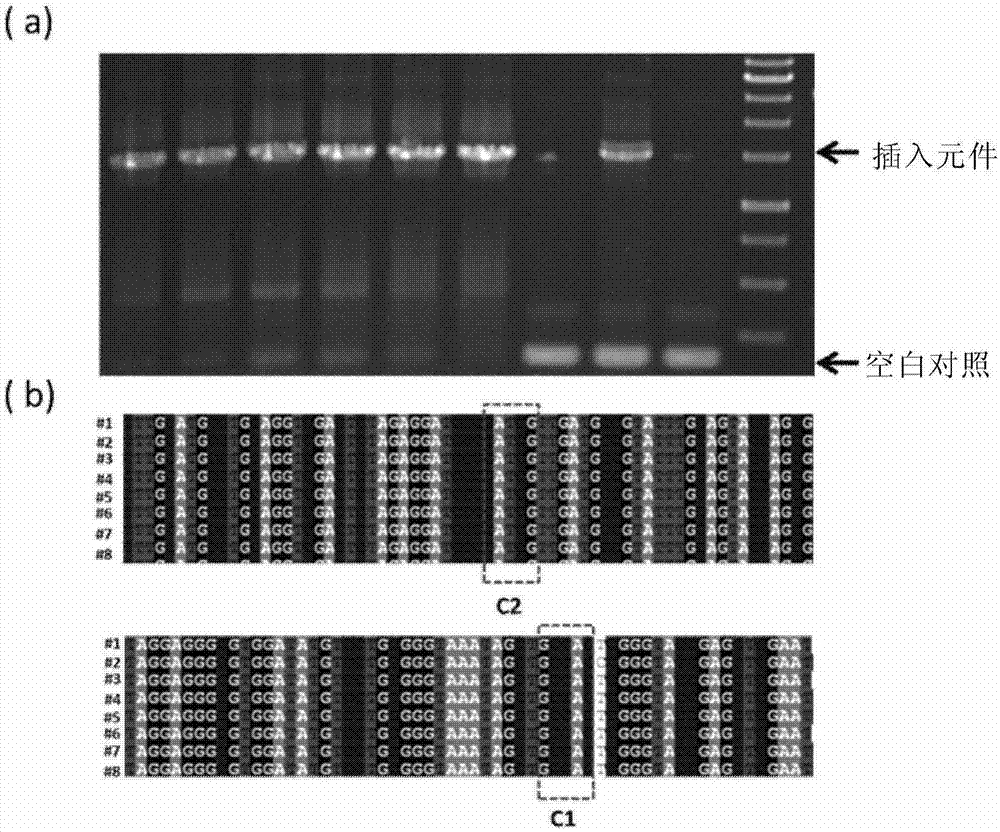
[0193] Example 1 The Apra resistance gene was seamlessly spliced and cloned into pUC18.
[0194] 1) using pBC-Am as a template, using primers apr-cf (SEQ ID NO.14) and apr-cr (SEQ ID NO.15) to PCR amplify the Apra resistance gene apr (SEQ ID NO.3), And 17-bp Cpf1 recognition sequence and PAM (TTN) were introduced at both ends by primers.
[0195] 2) Using pUC18 (SEQ ID No.2) as a template, use primers pUC18-cf (SEQ ID NO.12) and pUC18-cr (SEQ ID NO.13) to amplify the linear vector by PCR, and introduce the primers at both ends 17-bp Cpf1 recognition sequence and PAM (TTN) and 5-bp linker sequence to create the same cohesive ends as apr (SEQ ID NO. 3).
[0196] 3) Recover and purify the above PCR products, mix them together in an equimolar ratio, add FnCpf1 (SEQ ID No. 1), crRNA1 (SEQ ID No. 4), and T4 DNA ligase, and react at 30° C. for 1 h.
[0197] 4) The above reaction product was inactivated at 65°C for 20 minutes, immediately placed on ice for 5 minutes, and transform...
Embodiment 2
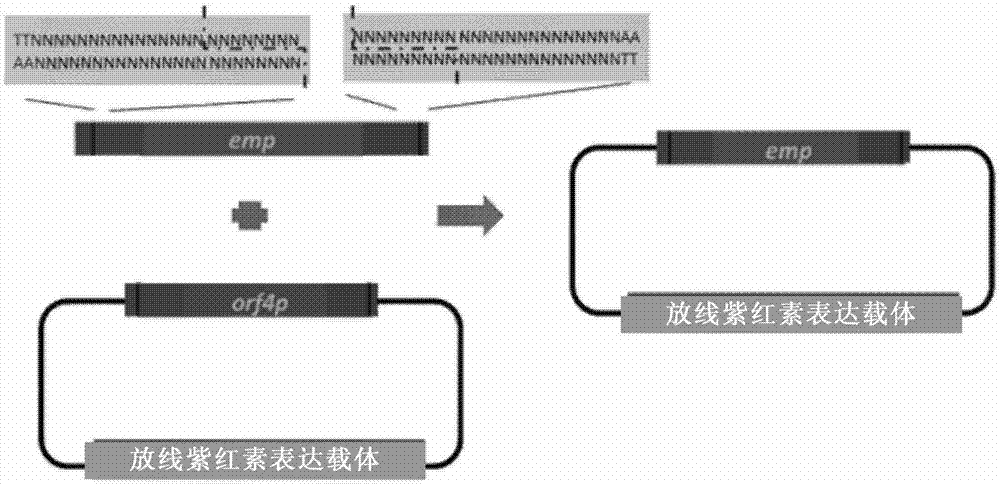
[0200] Example 2 The promoter of the pathway-specific regulatory factor actII-orf4 in the actinhodine biosynthetic gene cluster was replaced by the constitutively expressed erythromycin promoter.
specific Embodiment approach
[0201] like image 3 As shown, through the mediation of 17-nt crRNA1 and crRNA2, FnCpf1 forms cohesive ends that are adapted at both ends on the HIW plasmid and pEASY-emp plasmid respectively, and actⅡ-orf4 (SEQ ID No.7) The promoter was replaced with the erythromycin promoter (SEQ ID No.8). The specific implementation is as follows:
[0202] 1) Using pBS-emp as a template, use primers emp-pf (SEQ ID No.17) and emp-pr (SEQ ID No.18) PCR amplification to obtain the erythromycin promoter (emp) (SEQ ID No.8 ), and cloned into pEASY-blunt (SEQ ID No.10) to obtain the subclone pEASY-emp, which was verified by Sanger sequencing. PAM (TTN), a 17-bp FnCpf1 recognition sequence, and upstream and downstream homologous sequences before the element to be replaced and the recognition sequence were introduced at both ends by primers.
[0203] 2) Actinhodine expression plasmids pHIW (SEQ ID No.9) and pEASY-emp were digested with crRNA2 (SEQ ID No.5) and crRNA3 (SEQ ID No.6) mediated by Fn...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com