Rapid screening method applied to model animal zebrafish transgenosis
A zebrafish and transgenic technology, applied in the field of genetic engineering, can solve the problems of a lot of screening work, time-consuming, and the zebrafish cannot achieve the expected effect, etc., to improve the efficiency of targeted cleavage, the effect of gene expression is obvious, and the tracking and monitoring of the living body is easy. Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
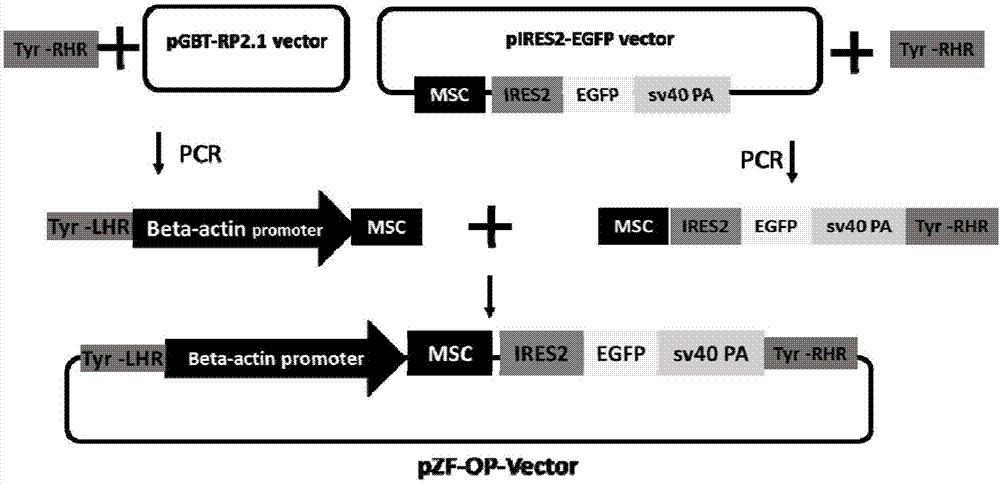
[0041] This Example 1 provides how to apply the method of the present invention to the method of targeted transfer into the zebrafish Nsun2 gene, and at the same time overexpress the zebrafish Nsun2 gene through a strong promoter. The transgenic homologous recombination vector pZF-Nsun2-OP-Vector containing the eGFP reporter gene was constructed, and the double-stranded gap mediated by CRISPR / Cas9 was used to insert it into the first exon region of the Tyrosinase gene by the MMEJ method to terminate the Tyrosinase gene. The expression of the Nsun2 gene was expressed at the same time, and finally the double-marker was used for high-efficiency screening to obtain the zebrafish individuals that were transferred to the Nsun2 gene and overexpressed the gene at the same time.
[0042] According to the difference of the transferred gene, the CDs region of the gene can be amplified and cloned, and constructed into the vector to achieve the purpose of transgenic. If the endogenous gene ...
Embodiment 2
[0092] This Example 2 provides the optimization and homology arm acquisition for the Tyrosinase gene target in the present invention.
[0093] Specifically include the following steps:
[0094] 1.1 Target design
[0095] Search zebrafish Tyrosinase (Danio rerio strain Tuebingen chromosome15.NC_007126.7) from NCBI, and predict the position of the target online at http: / / chopchop.cbu.uib.no / . Select the four highest scores as candidate target positions, and the target design mode is as follows: Figure 11 shown.
[0096] 1.2 Validation of targets
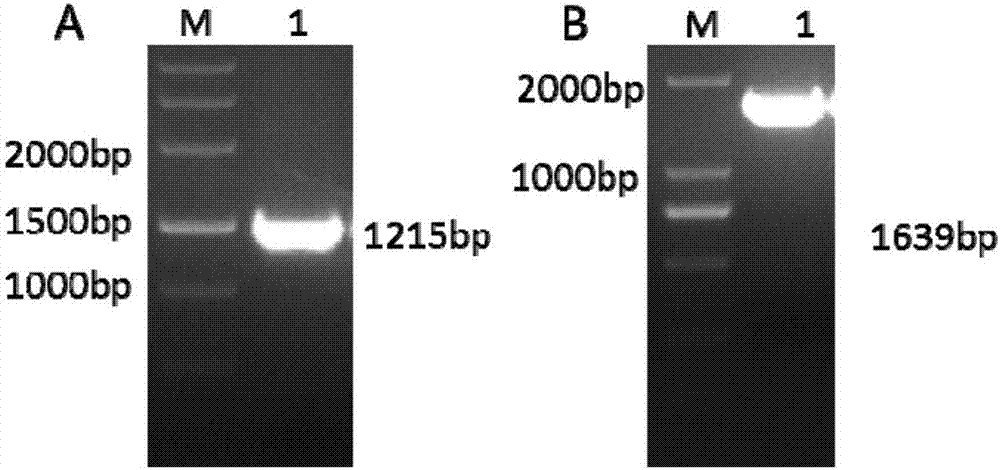
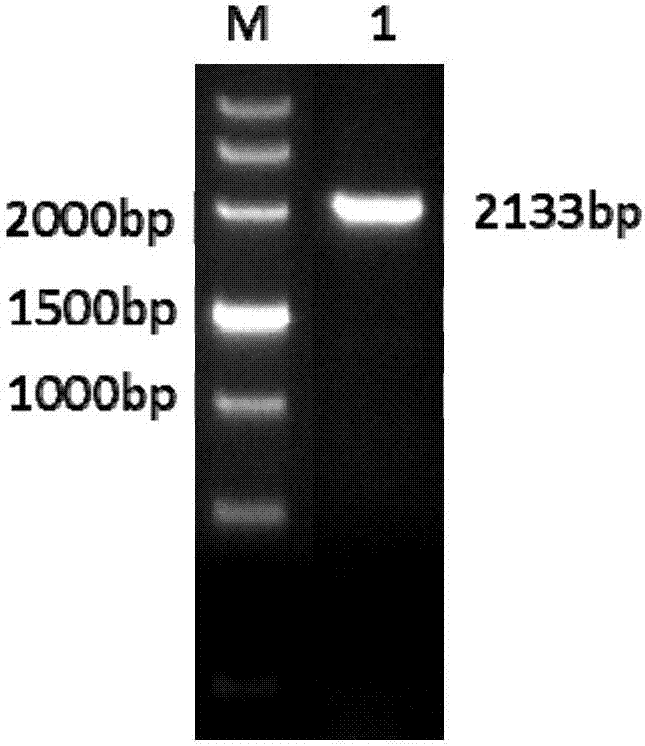
[0097] Add T7promoter to the upstream of the target site, add an overlap to the downstream, then synthesize primers and anneal to obtain PCR products. Using the PCR product as a template, transcribe in vitro to form sgRNA, co-inject zebrafish with Cas9-mRNA, and collect juvenile fish at 60hpf, and partially knockout juvenile fish such as Figure 12 As shown in A, DNA was extracted from juvenile fish of different groups. The targ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com