Pig-whole-genome low-density SNP chip and manufacturing method and application thereof
A technology of whole genome and production method, applied in the field of gene and molecular breeding, can solve the problems of difficult implementation and high cost of high-density chips, and achieve the effect of reducing costs and improving the breeding process.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
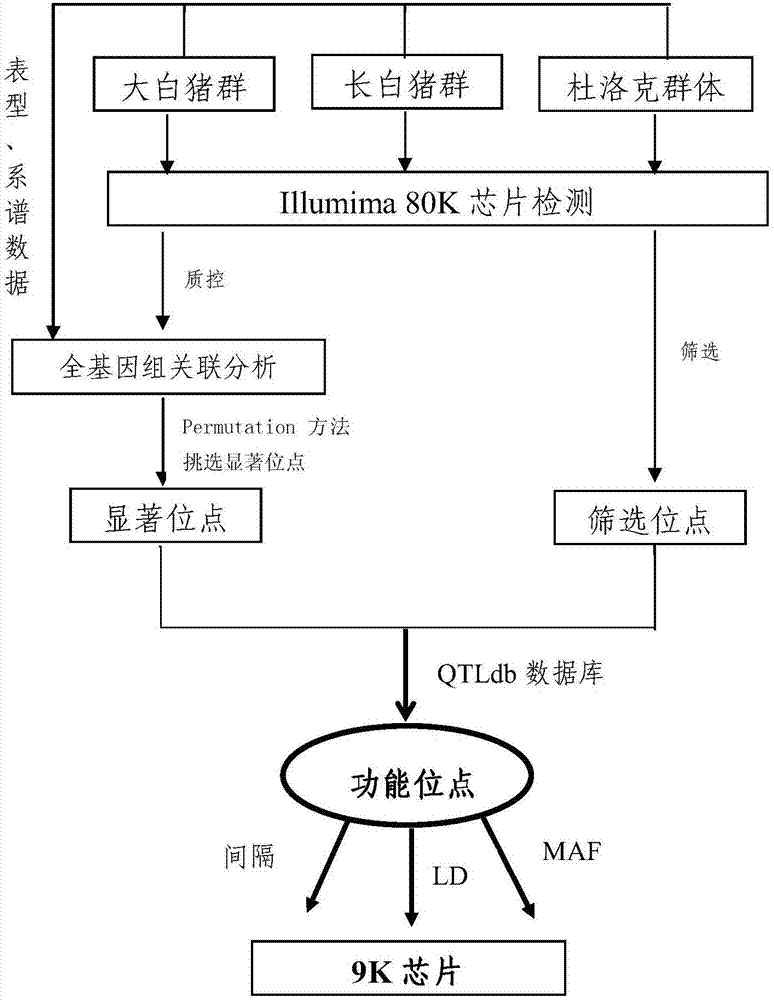
[0041] Example 1: Production of a low-density SNP chip for the whole pig genome
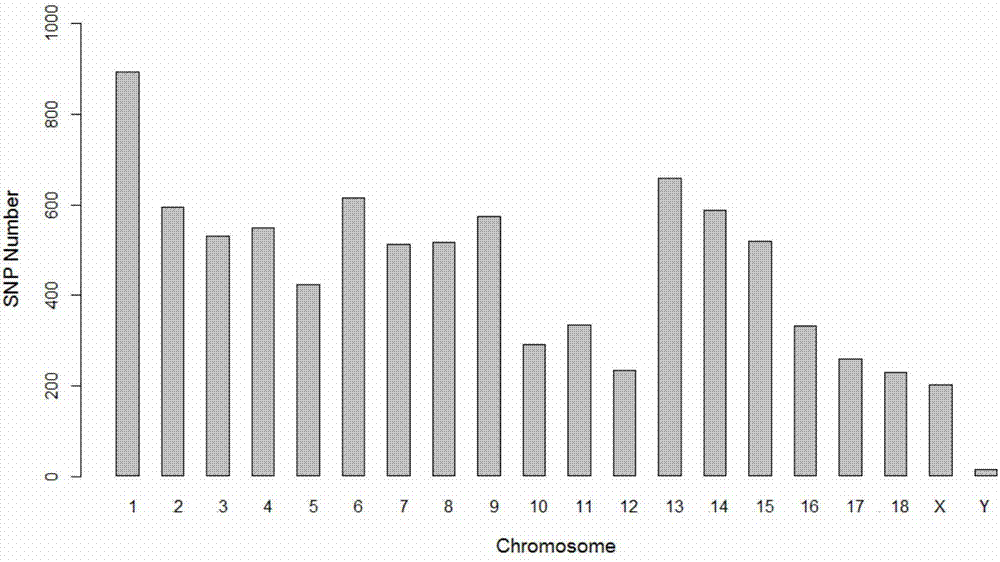
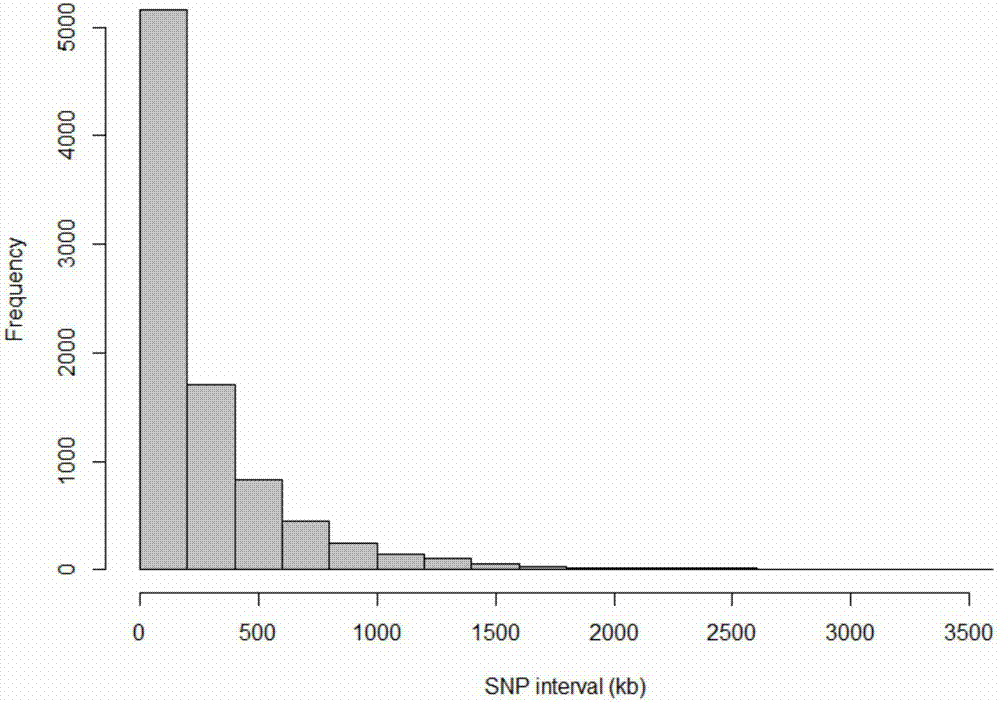
[0042] The pig whole genome low-density SNP chip of the present invention is referred to as the 9k chip for short. The 9K chip is mainly based on GGP-PorcineHD (68528SNP, developed by geneseek company, referred to as 80K). The degree of linkage disequilibrium between SNP markers has hardly decreased, 0.54 for 80K, and 0.53 for 9K.
[0043] The linkage disequilibrium of SNP markers is the guarantee of the accuracy of genome-wide association analysis and genome selection. The lower the linkage disequilibrium, the lower the accuracy of genome selection will be. At the same time, the allele frequency MAF of the 9K chip has little change with that of the 80K chip. These indicators show that the cost of 9K chips has decreased, but the use effect can still maintain a level close to that of 80K. See Table 1-1 for details.
[0044] Table 1-1 is the comparison of 9K and 80K SNP chip parameters
[0045] ...
Embodiment 2
[0252] Embodiment two: this implementation utilizes the designed 9K chip to estimate the accuracy of the genome breeding value, and compares it with the accuracy of the GGP-PorcineHD (68528SNP, referred to as 80K) chip, including the following steps:
[0253] (1) Obtaining data, the source of data is 27,081 large white pigs born in a pig farm in Beijing between 2007 and 2016. GGP-Porcine HD SNP chip genotype determination was performed on 1,429 sows, and 100 kg for 2 traits Traditional breeding value estimation and corrected phenotype calculations were performed based on age in days and number of piglets born alive. Select 1,429 heads for genotype determination for 9K chip design, and edit the SNP chip data, a total of 8,552 SNPs are used for analysis; select 1,159 of them as a reference group at the age of 100 kg, and the remaining 270 youngest heads as a verification group ; 411 heads were used as a reference group for the number of live litters, and the youngest 210 heads w...
Embodiment 3
[0258] Example 3: Using 9K chip information to identify the genetic relationship between populations
[0259] (1) Obtaining data, the samples come from 1156 American Large White pigs and 878 British Large White pigs from two different pig farms. First, use 26,122 pieces of pedigree information from the American large white and 31,802 pieces from the British large white to construct the A matrix based on the pedigree information, and construct the G matrix and the Kinship matrix based on the chip information with 1,156 American large whites and 878 British large whites from the 9K chip, and calculate the relationship between individuals. kinship relationship between. And use the G array to conduct paternity test on 1156 American large whites to evaluate the accuracy of the pedigree information.
[0260] (2) Use the pedigree information to construct the A array, that is, the molecular relationship matrix (numerator relationship matrix, NRM), and use the 9K chip information to c...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com