Primer for inhibiting sequence-specific amplification of 18S rDNA of shrimps and oysters and application thereof
A specific, oyster technology, applied in the field of PCR primers, to achieve the effect of convenient research
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
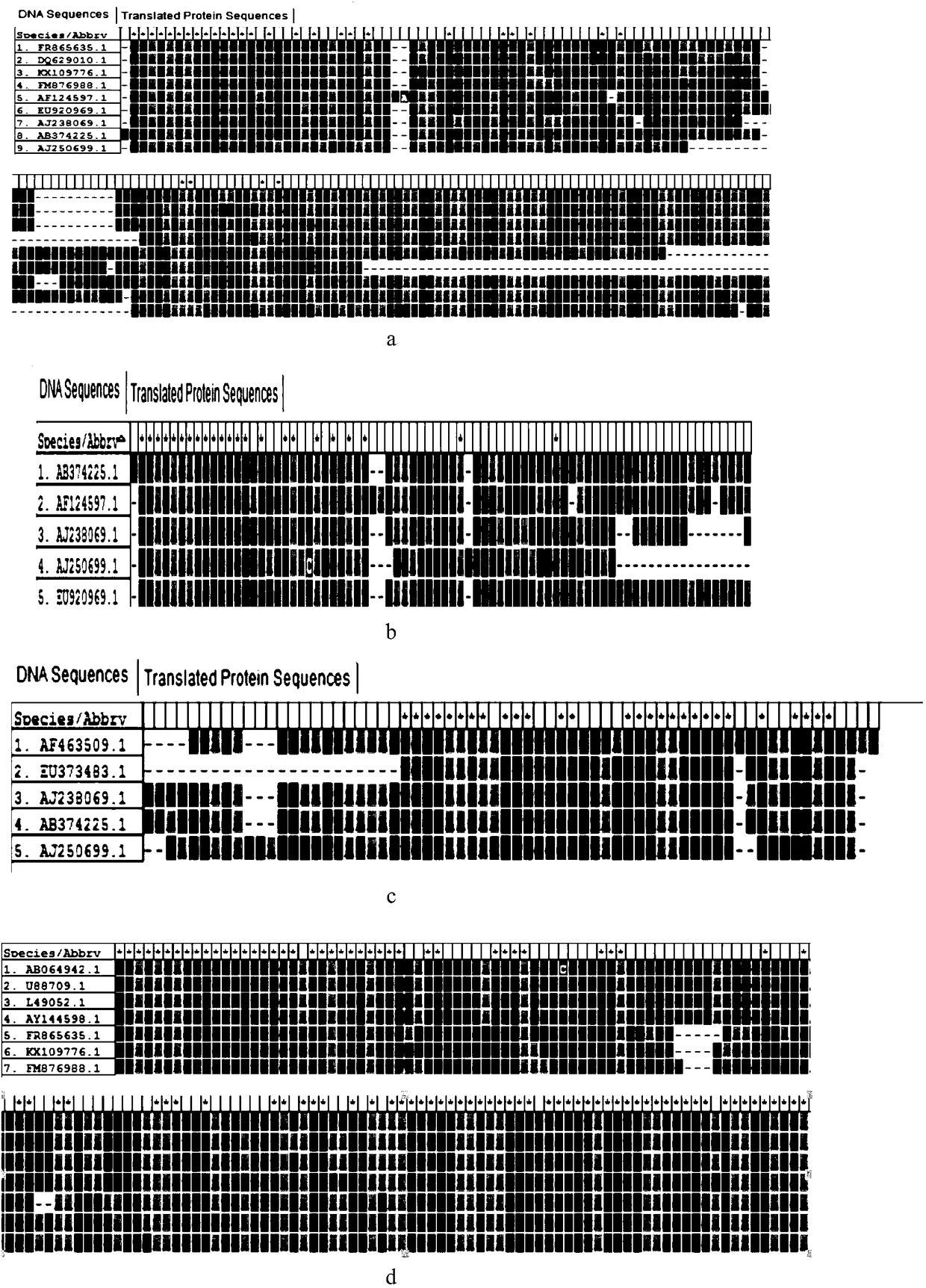
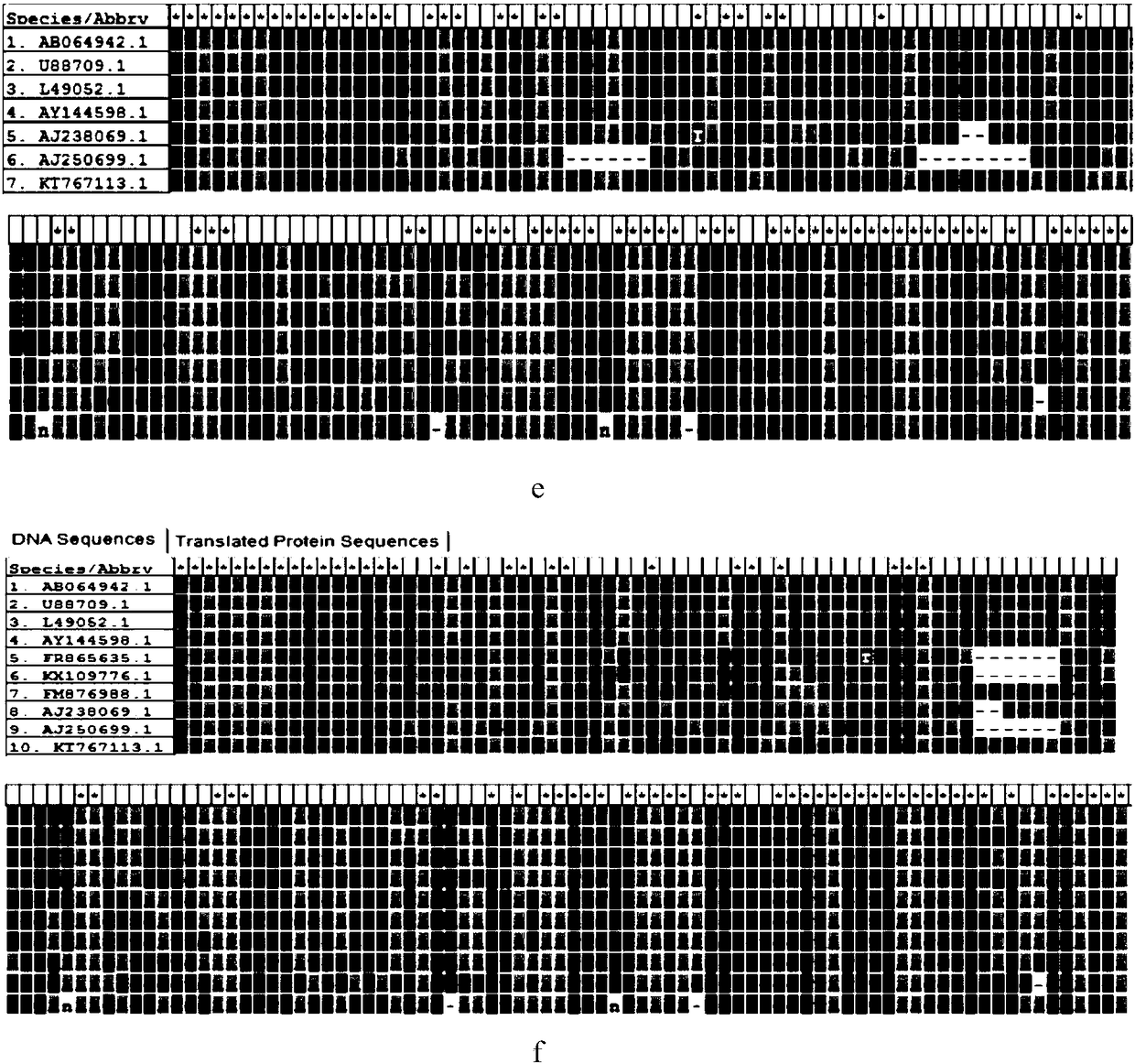
[0023] Example 1 Design of inhibitory primers
[0024] Suppressing primer design scheme:
[0025] 1. Design of C3 spacer
[0026] There is a certain overlap between the 5'end (18-25nt) and the F(R) 3'end of the universal primer. The subsequent sequence should be as consistent as possible with the 18S gene sequence to be suppressed, and different from the gene sequence that is not desired to be suppressed, and finally at the end Add C3spacer.
[0027] 2. Design of dual-priming oligonucleotides
[0028] The design of the 5'end is similar to the 5'end of C3, except that 44444 (4=dlnosine) is added before the 3'end (6-12nt), and finally C3spacer is added to the end.
[0029] The design process of suppression primers:
[0030] Such as figure 1 As shown, according to Penaeus vannamei (GenBank accession no.EU920969.1), Penaeus vannamei (GenBank accession no.EU118282.1), Penaeus vannamei (GenBank accession no.AF186250.1), Penaeus vannamei (GenBank accession no. AF124597.1), Penaeus vannamei (Ge...
Embodiment 2
[0035] Example 2 Verification of suppression primers
[0036] The DNA of Penaeus vannamei and Hong Kong oyster were extracted using the Mollusk DNA Extraction Kit (Magen, Suzhou, China), and the DNA of plankton microorganisms and zooplankton in the water filtered onto the membrane were extracted using the Water DNA Extraction Kit (Magen, Suzhou, China) ) The specific operation is carried out in accordance with the instructions on the manual.
[0037] For the inhibitory primer nmb-BP2-DPO, configure the inhibition reaction system with different concentrations of 0.01μM, 0.02μM, 0.04μM, and use ddH 2 O was used as a negative control, and the reaction without inhibitory primers was used as a positive control. The templates used the digestive tract tissue DNA of Penaeus vannamei, the digestive tract tissue DNA of Hong Kong oyster, the planktonic microorganism DNA filtered to the membrane, and the zooplankton DNA. Set up 3 test replicates for different template DNAs, and amplify them by...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com