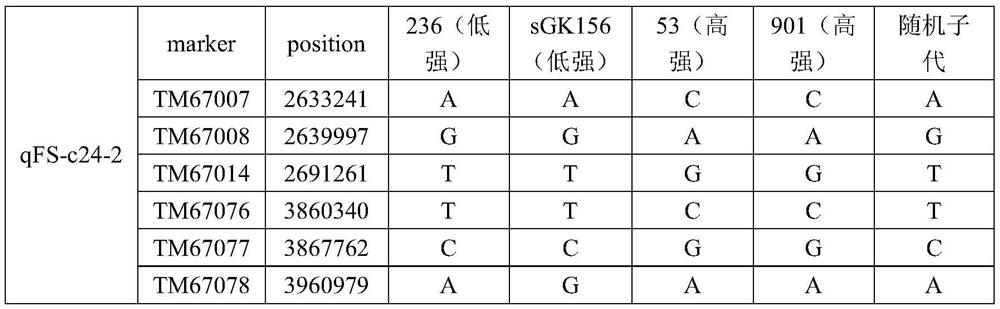
Mapping of qtl, SNP molecular markers and related genes related to fiber strength on chromosome 24 of 70 population of China Cotton Research Institute
A technology of molecular markers and cotton fibers, which is applied in the determination/inspection of microorganisms, recombinant DNA technology, DNA/RNA fragments, etc., and can solve problems such as large distances, inability to locate genes, and inaccurate physical locations
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0022] 1. Obtaining recombinant inbred line population
[0023] Using upland cotton lines SGK156 and 901-001 as parents, the parents were planted in the experimental base of the Cotton Research Institute of the Chinese Academy of Agricultural Sciences in Anyang City, Henan Province in 2011, and the F 1 , named as the variety Zhongmiansuo 70, F 1 The plants were self-pollinated, and the combination was propagated in the south of Sanya City, Hainan Province in the winter of 2011, and 250 F 2 Single plant F 2:1 , and then in 2012, 250 individual plants were self-planted in a row to obtain F 2:3 , planted 250 rows of F in Anyang in April 2013 2:3 Strains, in September of the same year, select 10 F plants with better performance in each row 2:1 After mixed harvest, go to Hainan Nanfanjiadai to get F 2:5 , and continued to plant F in Anyang in 2013 2 Single plant, continue to plant F in Anyang in 2014 2:5 Plant line. from F 2:3 Starting from the strains, the planting norms ...
PUM
| Property | Measurement | Unit |
|---|---|---|
| hardness | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com