Method for evaluating X protein binding molecule of human hepatitis C virus
A hepatitis B virus, binding molecule technology, applied in the fields of biotechnology and genetic engineering, can solve the problems of difficult high-throughput detection research, time-consuming and labor-intensive, and high technical requirements for operators, achieving signal sensitivity, meeting screening and evaluation requirements Requirements, the effect of convenient detection
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0042] Embodiment 1: Construction of recombinant fusion protein expression vector
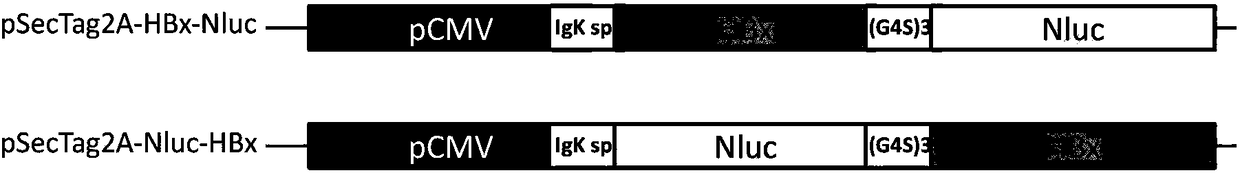
[0043] Construct pSecTag2A-HBx-Nluc expression vector:
[0044] Use the primer pair HBx_F1 / HBx_R1 to amplify the HBx gene sequence, and recover the PCR reaction fragment; use the primer Nluc_F1 / Nluc_R1 to amplify the Nluc gene sequence, and recover the PCR reaction fragment; use the primer HBx_F1 / Nluc_R1 to perform Overlap PCR on the HBx and Nluc gene fragments, and get The fragment of the PCR product was cloned into the secretory eukaryotic expression vector pSecTag2A (purchased from Invitrogen, the vector has a nucleic acid sequence encoding the Igκ signal peptide), to obtain the expression vector pSecTag2A-HBx-Nluc of the fusion gene HBx-Nluc (such as figure 1 above);
[0045] Construct pSecTag2A-Nluc-HBx expression vector:
[0046] Use primers Nluc_F2 / Nluc_R2 to amplify the Nluc gene sequence, and recover the PCR reaction fragment; use the primer pair HBx_F2 / HBx_R2 to amplify the HBx gene...
Embodiment 2
[0047] Example 2: Expression and detection of fusion protein
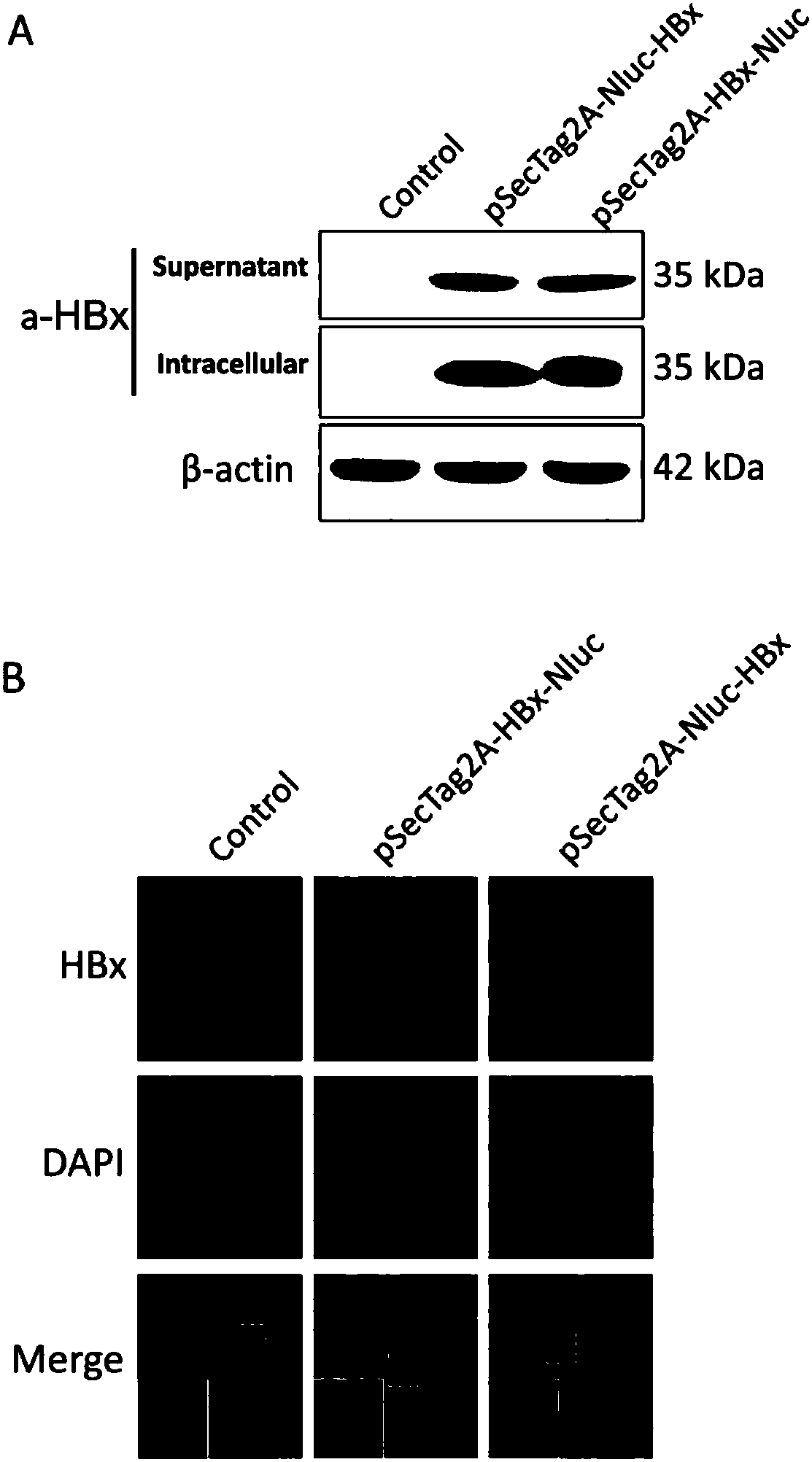
[0048] The fusion protein expression plasmid was transiently transfected into Huh7 cells, and after a certain period of time, the expression of the fusion protein was detected using the specific antibody of HBx protein;
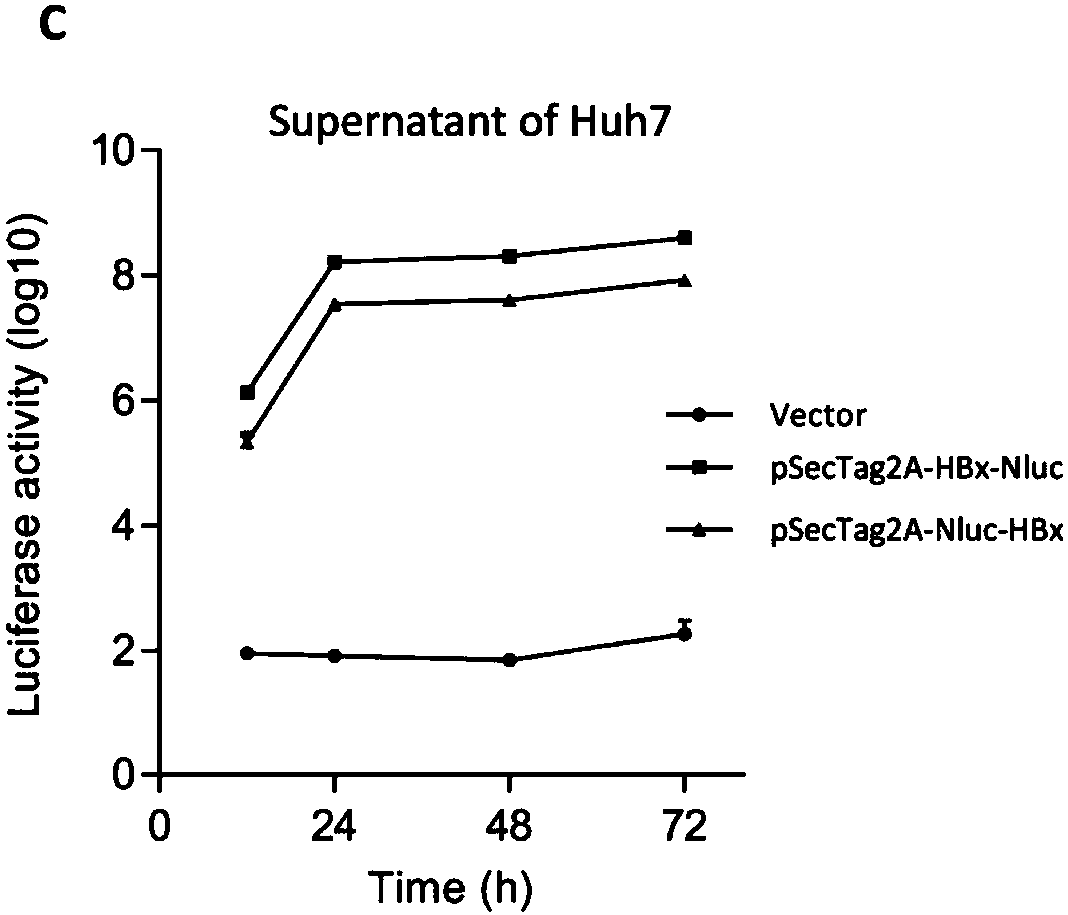
[0049] Western blot results show that the fusion protein can be recognized by the specific antibody of HBx protein, and its molecular weight is about 35kDa (such as figure 2 Shown in A), the results of immunofluorescence showed that the fusion protein was distributed in both the nucleus and the cytoplasm (such as figure 2 Shown in B), as time goes by, the activity of the luciferase detected in the supernatant gradually increases, and reaches a relatively high expression level in 48 hours (as figure 2 C shown).
Embodiment 3
[0050] Embodiment 3: RNA interference suppression experiment
[0051] Use shRNA targeting HBx gene to inhibit the expression of fusion protein in cells (such as image 3 shown), the positive control showed that the shRNA targeting the HBx gene could knock down the expression of the HBx gene (such as image 3 Shown in A); this shRNA can also knock down the expression of fusion protein (such as image 3 Shown in B); along with the reduction of fusion protein expression, the Nluc luciferase activity in the cell supernatant correspondingly significantly reduces (as image 3 C shown).
PUM
| Property | Measurement | Unit |
|---|---|---|
| molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com