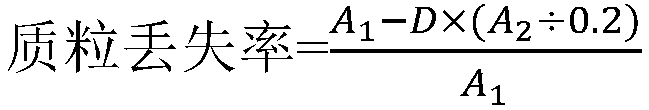Yeast genome editing carrier as well as construction method and applications thereof
A technology of genome editing and construction methods, applied in other methods of inserting foreign genetic materials, genetic engineering, biochemical equipment and methods, etc., can solve the problem that the positive clone rate of multi-site genome editing is not high, and strains cannot be edited again , Multi-site editing efficiency is low at the same time, to achieve the effect of multiple rounds of genome editing
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
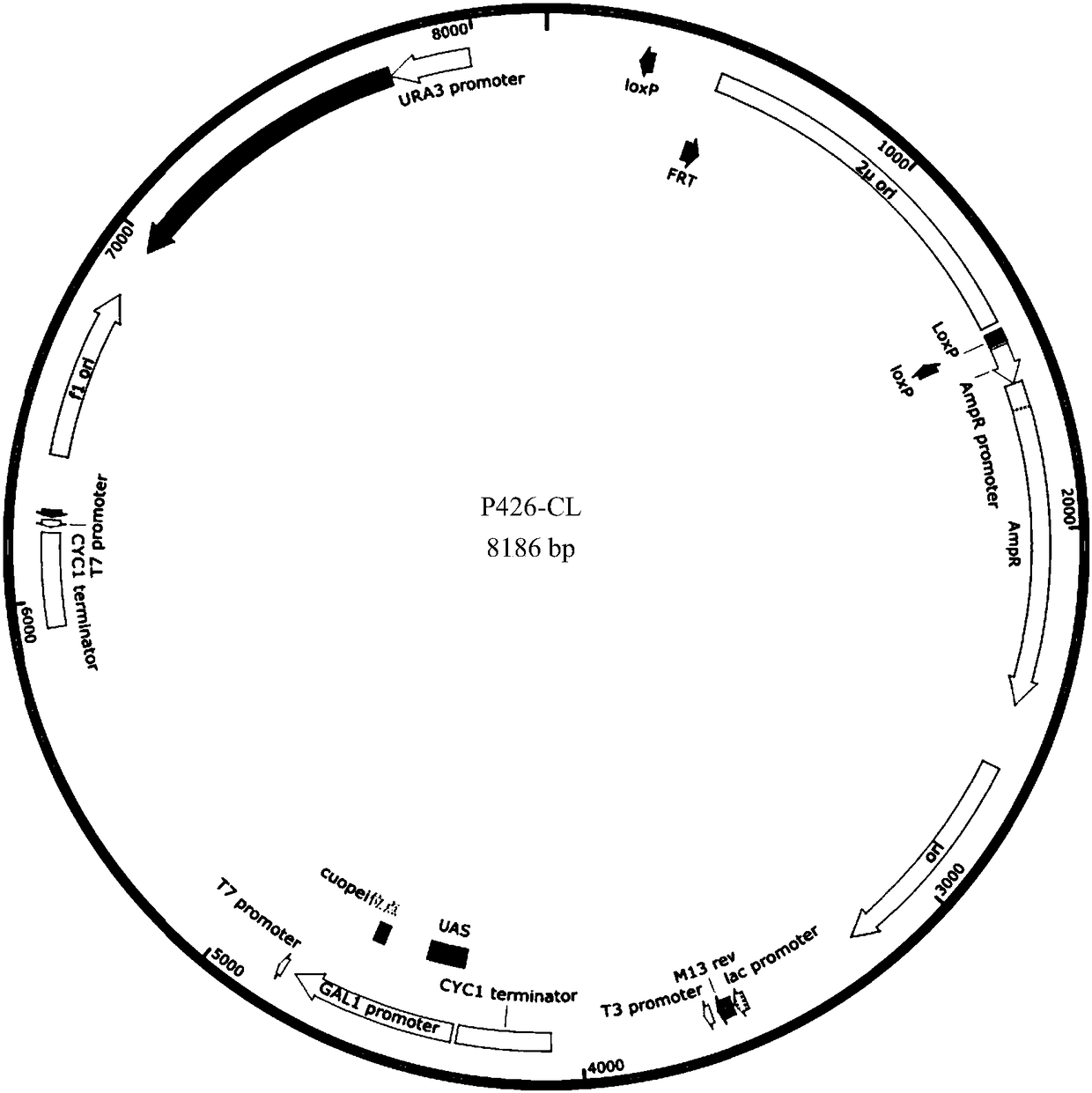
[0026] Example 1 Construction of Yeast Genome Editing Universal Vector
[0027] Using Addgene's commercialized plasmid p426-SNR52p-gRNA.CAN1.Y-SUP4t (referred to as P426) as the starting vector, the same LoxP sequence (SEQ ID NO: 1) was introduced at both ends of the Ori site of the vector, and at P426 The KpnI site is integrated into the complete recombinase Cre expression cassette. The specific implementation is as follows:
[0028] 1. Introduction of LoxP sequences at both ends of the replication origin site Ori
[0029] Using the P426 vector as a template, the LoxP1 fragment was amplified with the P1 (5'-TCGTATAGCATACATTATACGAAGTTATACTTATATGCGTCTATTTATGT AG-3') / P2(5'-GTATAGCATACATTATACGAAGTTATTCCCCCGAAAAGTGCCACCTGA-3') primer pair. II Recombination Cloning Kit (Nanjing Novizan Biotechnology Co., Ltd.), the plasmid P426-LoxP1 was obtained by recombining and circularizing LoxP1 itself.
[0030] Further, using P426-LoxP1 as a template, the LoxP2 fragment was obtained by a...
Embodiment 2
[0038] use II recombination cloning kit will PCR product gene fragment P GAL1 -Cre-T CYC1 Homologous recombination with KpnI linearized P426-LoxP to obtain the yeast genome editing universal vector p426-CL. Example 2 Knockout of Geranylgeranyl pyrophosphate synthase (Geranylgeranyl pyrophosphate synthase, BTS1) and transcriptional regulatory gene (Heme-dependent repressor of hypoxic genes, ROX1) gene in Saccharomyces cerevisiae genome
[0039] The vector P426-CL constructed in Example 1 was used as the gRNA expression vector to construct the backbone to realize the knockout of the BTS1 and ROX1 genes in the yeast genome.
[0040] 1. PCR amplification of bts1-gRNA and rox1-gRNA expression vector fragments
[0041] Using P426-CL as a template, P13(5′-TAATAATGGTTTCTAGTATGA-3′) / P14(5′-AACAGGATCATTATTGATCAGATCATTTATTCTTACTGC-3′) and P15(5′-TGATCAATAATGATCCTGTTGTTTTGAGCTAGAAATA-3′) / P16(5′-ACTAAGAAAACCATTATTATCAT-3 ') is a pair of primers, respectively amplified to obtain fragme...
Embodiment 3
[0054] Example 3 Saccharomyces cerevisiae S.cerevisiae BJ5464ΔBTS1ΔROX1 genome ERG9 gene, ypl062w gene and yjl064w gene editing
[0055] The P426-CL constructed in Example 1 was used as a template as a gRNA expression vector to construct the backbone to realize the editing of the yeast genome ERG9 gene, yp1062w gene and yjl064w gene.
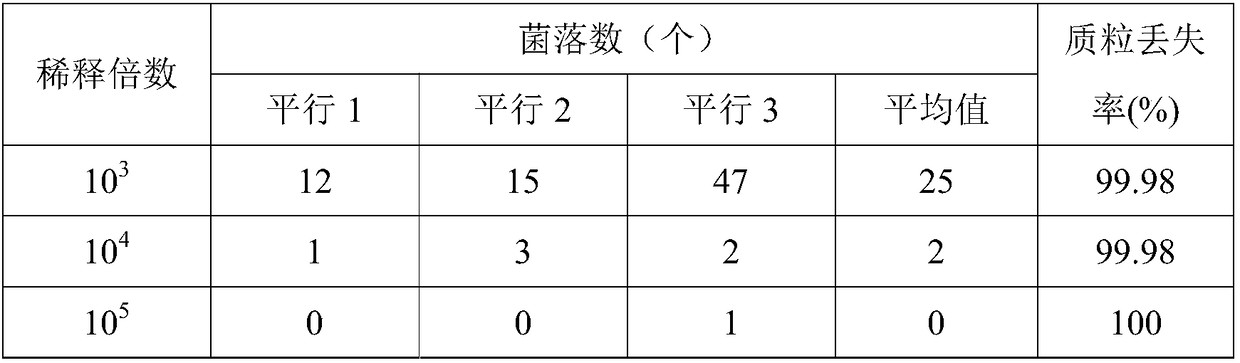
[0056] 1. Loss of carrier P426-CL-BTS1-ROX1 in S. cerevisiae BJ5464ΔBTS1ΔROX1 / P426-CL-BTS1-ROX1 cells
[0057] Pick a single colony of S.cerevisiae BJ5464ΔBTS1ΔROX1 / P426-CL-BTS1-ROX1 and inoculate it in SG / ΔTrp liquid medium (galactose 20.0g / L, DO Supplement 0.62g / L, amino-free yeast nitrogen source YNB 6.7g / L , Ura 0.02g / L, Leu0.06g / L), cultivated at 30°C and 220rpm for 36h. Dilute the bacterial solution to 10 6 After doubling and spread on SD / ΔTrp solid plate containing 1g / L 5-FOA (glucose 20.0g / L, agar 20.0g / L, DO Supplement 0.62g / L, amino-free yeast nitrogen source YNB 6.7g / L , Ura 0.02g / L, Leu 0.06g / L), cultivated at 30°C for 36h, passed...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com