Homocysteine methyltransferase mutant and application thereof, as well as nucleic acid, expression vector, host cell, reagent
A cystine methyl, host cell technology, applied in homocysteine methyltransferase mutants and their applications, as well as in the fields of nucleic acids, expression vectors, host cells, and reagents, can solve the problem of low activity and low relative activity and other problems, to achieve the effect of high expression, stable recombinant plasmid and improved stability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0051] Example 1 Error-prone PCR (error-prone PCR) method to construct HMT mutation library
[0052] use Gene The Morph II random mutagenesis kit uses the optimized nucleotide sequence of HMT (see SEQ ID NO.2) as a template to amplify the HMT gene and introduce mutations randomly.
[0053] Amplification primer
[0054] F: 5'-CATG CCATGG CTAGATTGCCATTGAAG (NcoI endonuclease is underlined)
[0055] R: 5'-CCG CTCGAG AGTGTACTTCTTAACAGCAG (XhoI endonuclease underlined)
[0056] Reaction conditions: The reaction conditions are: pre-denaturation at 94°C for 10 minutes, denaturation at 94°C for 30 seconds, annealing at 60°C for 60 seconds and extension at 72°C for 2 minutes, a total of 25 cycles, 0.8% agarose electrophoresis, and the kit to recover the target gene fragment . According to the method described in the product manual of NEB Company, after double digestion with NcoI and XhoI, ligate with the pET-28a(+) vector (kana resistance) that has been digested with NcoI and X...
Embodiment 2
[0057] Example 2 Screening of HMT mutant library
[0058] After the mutant library clones in Example 1 were collected, the plasmids were extracted, transformed into Escherichia coli expression strain BL21 (DE3), spread on LB plates containing kana, and cultured for 12 hours. Pick a single clone in a 96-well plate, each well contains 150 μL of TB medium (containing 50 μg / mL kanamycin, 1 mM IPTG), 37 ° C, 245 rpm, shaking culture for 36 h. The 96-well plate replicator replicated each single clone on an LB solid medium plate, cultured at 37°C for 12 hours, and stored in a refrigerator at 4°C. Gently suck out the cell cultures in each well of the 96-well plate with a row gun, and distribute them in the 96-well plates of plate A and plate B according to the corresponding positions, and the culture in each well of each 96-well plate is 70 μL. Centrifuge at 4000rpm at 4°C for 10min, discard the supernatant, and resuspend the bacterial cells in each well with 30μL, 50mM, pH7.6 sodium...
Embodiment 3
[0059] Embodiment 3 Stability detection
[0060] The mutants in plate B corresponding to the increased activity in plate A were tested for stability.
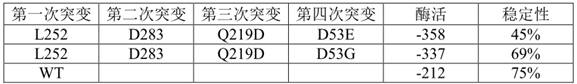
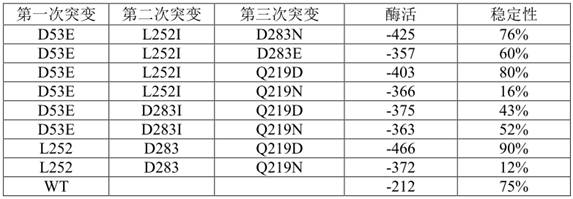
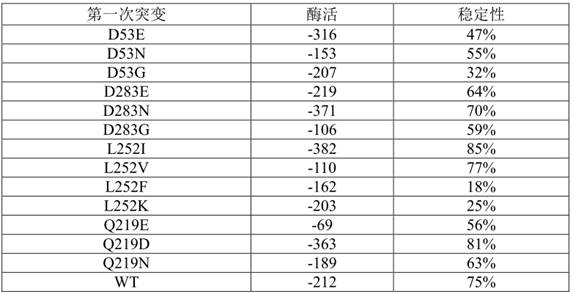
[0061] Screen out 4 mutants that affect the activity, and send the mutant monoclonal to the sequencing company for sequencing. The amino acid sequence is shown in SEQ ID NO.3-SEQ ID NO.6
[0062] The four mutation positions are D53, D283, L252, and Q219, respectively.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com