Method for rapidly identifying charybdis feriata sex specific molecular marker and genetic sex
A molecular marker, a technology for the rust-spotted cricket, which is applied in the directions of biochemical equipment and methods, microbial determination/inspection, DNA/RNA fragments, etc. problem, to achieve the effect of simple operation, short time-consuming, high identification accuracy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
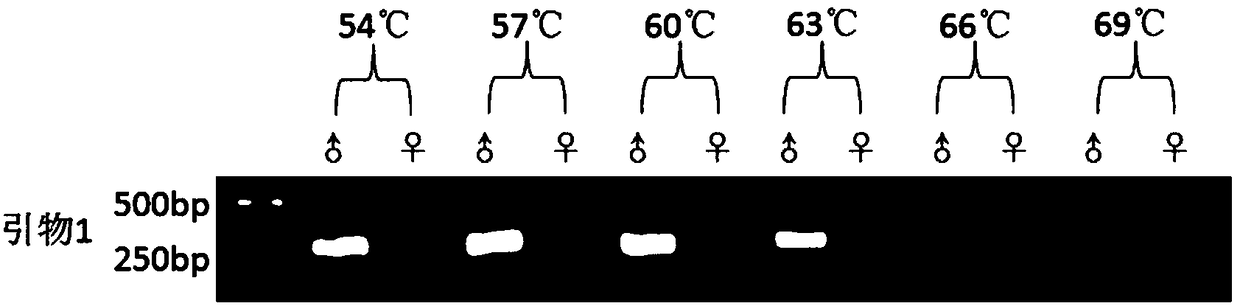
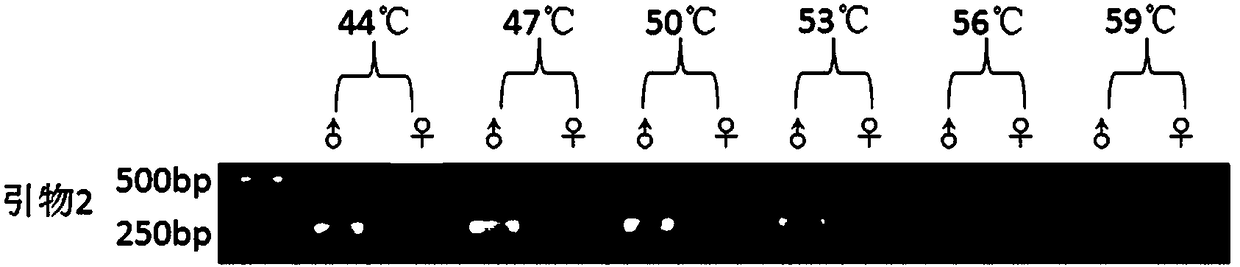
[0043] A method for quickly identifying sex-specific molecular markers and hereditary sex of rusty spot moth, mainly comprising the following steps:
[0044] 1. Sample collection and extraction of genomic DNA
[0045] A total of 44 samples were collected, including 22 males and 22 females. Take about 50 mg of the muscle tissue of P. rust, put it into a 1.5ml centrifuge tube containing 300 μL of lysate, cut it into pieces until it is homogenized, add 10 μL of RNaseA, mix well, and incubate at room temperature for 2 minutes. Then add 300 μL of lysate and 5 μL of proteinase K to the centrifuge tube, mix thoroughly, and put it in a 55°C water bath for 2 hours for digestion. Then add 300 μL Tris-balanced phenol and 300 μL chloroform for extraction, and centrifuge at 13000r for 10 minutes after 10 minutes. Draw about 500 μL of centrifuged supernatant into a new centrifuge tube, add 600 μL of chloroform to it and extract again. Draw about 350 μL of the supernatant after centrifuga...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com