Probe for in situ nondestructive testing of nucleic acid molecules in extracellular vesicles and preparation method and application thereof
A non-destructive detection and nucleic acid molecular technology, applied in the field of medical detection, can solve the problems of inaccurate detection of EVsmiRNA, lack of protection for the integrity of extracellular vesicles, and leakage of EVs contents, so as to improve detection and screening efficiency and save energy. The effect of shortening the sample size and detection time
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
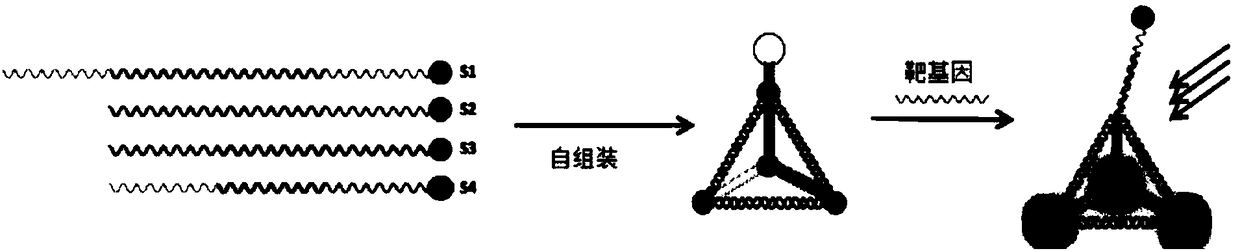
[0032] Example 1. Preparation of tetrahedral probe
[0033] Synthesize 4 single-stranded nucleotide sequences, as follows:
[0034] The first chain: (SEQ ID NO.1);
[0035] The second chain: (SEQ ID NO. 2);
[0036] The third chain: (SEQ ID NO.3);
[0037] Fourth chain: (SEQ ID NO.4);
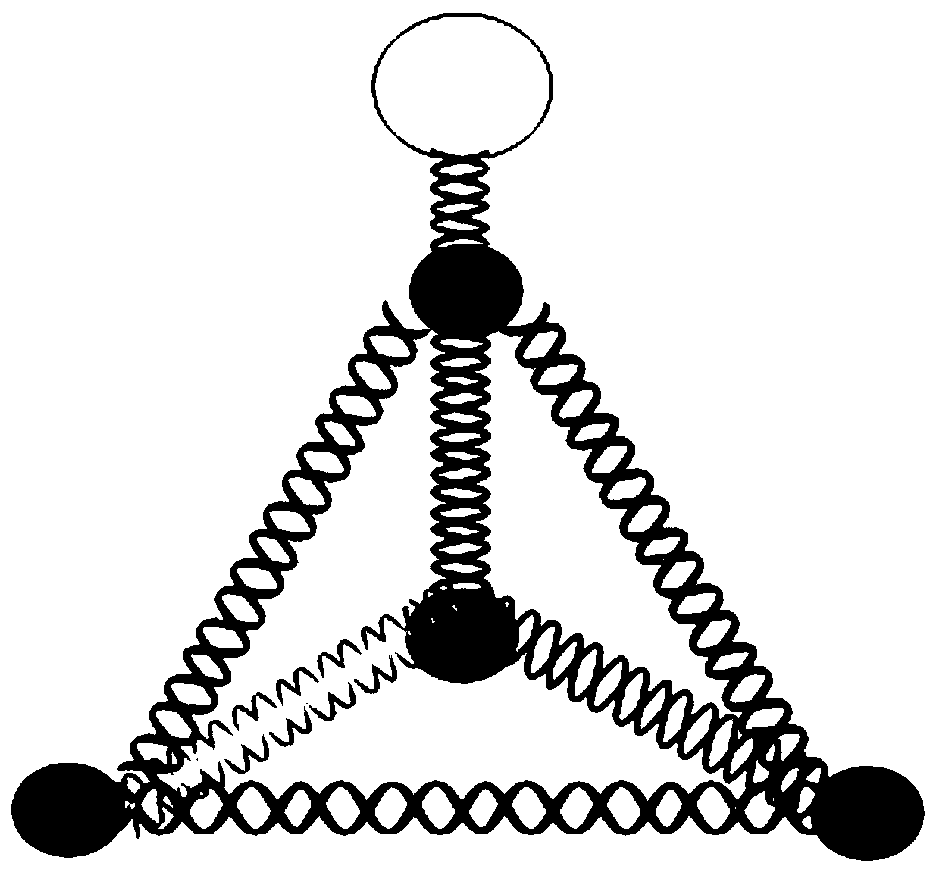
[0038] The complementary part of each chain in the above sequence uses the same label. The length of the complementary sequence is 13bp, the non-complementary bases are marked in black, the interval between complementary sequences is 2nt, the front of the chain is 1nt, and the bold in the first strand has no complement. Sequence to form a stem-loop probe.
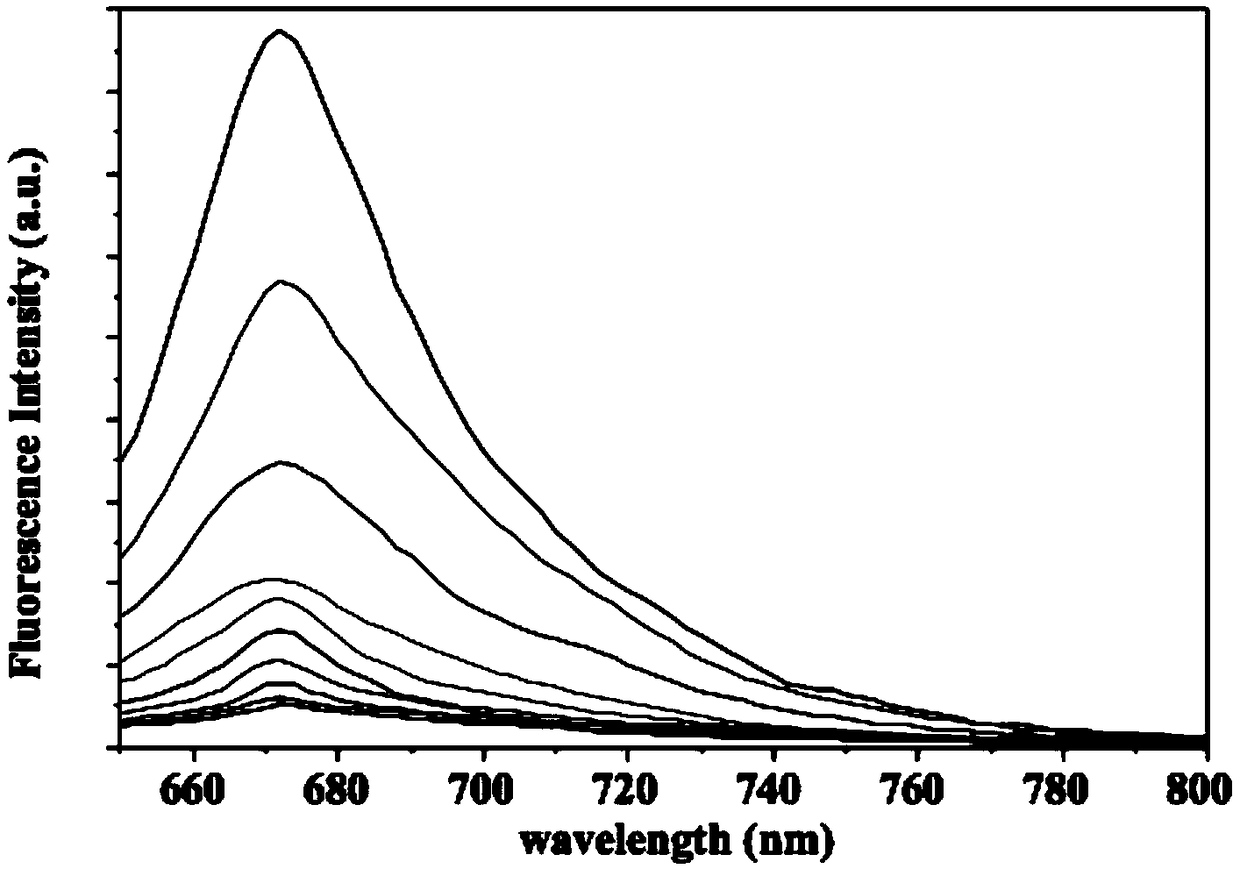
[0039] In order to be able to detect by fluorescence during detection, a fluorescent group was modified at the 5'end of the second, third and fourth chains, and a quenching group was modified at the 5'end of the first chain.
[0040] Dissolve the synthesized single chain in a single-strand self-assembled ion buffer. The preparation method of the sin...
Embodiment 2
[0048] Example 2. Preparation of hexahedral probe
[0049] The nucleotide sequence of synthetically assembled hexahedral probe is as follows:
[0050] First chain 1: (SEQ ID NO.5);
[0051] Second chain 2: (SEQ ID NO.6)
[0052] Third chain 3: (SEQ ID NO.7)
[0053] Fourth chain 4: (SEQ ID NO. 8)
[0054] Fifth chain 5: SEQID NO.9)
[0055] Sixth chain 6: SEQID NO.10)
[0056] The complementary parts of each chain in the six chains use the same label. The complementary length of the hexahedron is 20bp, and the non-complementary bases are black marked with 1nt; the fluorescent group is modified at the 5'of the first, second, third, and fourth chain. At the end, each chain is modified with a fluorescent group; the quenching group is modified at the 5'end of the 5th and 6th chain, and each chain is modified with a quenching group.
[0057] Then the synthesized single strands were self-assembled according to the method in Example 1 to form a hexahedral probe with the structure as Figure...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com