CRISPR/Cas9-gRNA targeting sequence pair, plasmid and HD cell model of HTT
A cas9-gRNA, cell model technology, applied in the field of plasmids and HD cell models, can solve problems such as practical application limitations, and achieve the effect of good gene editing activity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0037] According to GenBank collection of mouse (C57BL / 6J) HTT gene information:
[0038] Description: huntingtin[Source:MGI Symbol; Acc:MGI:96067]
[0039] Synonyms: IT15,C430023I11Rik,Hdh,HD,htt,huntingtin
[0040] Location: Chromosome 5:34,761,740-34,912,534forward strand.
[0041] Gene ID: 15194
[0042] And comprehensively analyze the HTT genome structure and protein function conserved region, design CRISPR target (gRNA), the target sequence is as follows:
[0043] HTT-L7: tccgccggcatgacgtcacggg;
[0044] HTT-L8: ccgcgagggttgccgggacgg;
[0045] HTT-L9: caagatggctgagcgccttgg;
[0046] HTT-L10:cattgccttgctgctaagtgg;
[0047] HTT-R7: ttggccatgcccagcacgcagg;
[0048] reverse complement: cctgcgtgctgggcatggccaa;
[0049] HTT-R8: taccgcgaccctctggacaggg;
[0050] reverse complement: ccctgtccagagggtcgcggta;
[0051] HTT-R9: cgggaaagcctggcctcaggg;
[0052] reverse complement: ccctgaggccaggctttcccg;
[0053] HTT-R10: cggctctgtctcctctgaggg;
[0054] Reverse complement: c...
Embodiment 2
[0090] This example provides the CRISPR / Cas9-gRNA targeting plasmid of HTT and its construction method. The targeting plasmid is obtained by constructing a pair of targeting sequences into a carrier plasmid. The targeting sequence pair is composed of L sequence and R sequence, so that the sense strand and antisense strand of the target gene (dsDNA) can be cut at the same time, so that the target gene can be knocked into the target site by HR. According to the in vitro activity detection results in Example 1, the activities of the Htt-L9 and Htt-R9 targets are both about 95%, indicating that the targeting plasmid composed of the Htt-L9 and Htt-R9 target pairing has a relatively high in vivo gene editing activity , so in this example, Htt-L9 and Htt-R9 were selected to form a targeting sequence pair, and the targeting sequence pair was constructed into a vector plasmid. The carrier plasmid is a carrier plasmid suitable for mammals and / or mammalian cells. In order to facilitate t...
Embodiment 3
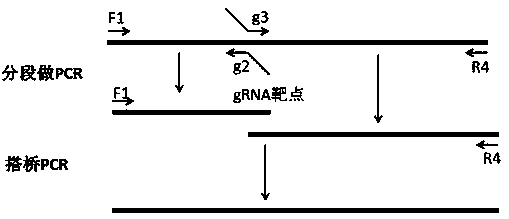
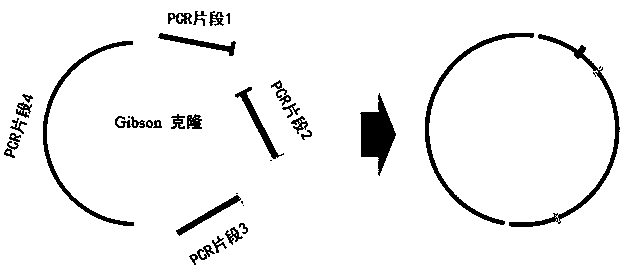
[0094] In this example, the NQ (150Q, 90Q, 50Q, 20Q) Donor plasmid was designed and constructed by Gibson cloning based on the nucleotide sequences and relative positions of the L and R targets of the Htt9 plasmid. Schematic diagram of Donor plasmid design and construction image 3 shown. image 3 In the figure, the restriction sites between PCR fragment 4 and PCR fragments 1 and 3 are Not1 restriction sites (represented in yellow in the original figure), and the restriction sites between PCR fragment 1 and PCR fragment 2 are EcoR1 and Xma1 restriction sites point (represented in red in the original figure), between PCR fragment 2 and PCR fragment 3 is the FRT (directed recombinase recognition site) site (represented in blue in the original figure).
[0095] 1. Extract CATH-a cell genomic DNA
[0096] CATH-a (ATCC CRL-11179) was purchased from Beijing Bena Chuanglian Biotechnology Research Institute and was frozen for this experiment. The cryopreserved cells were resuscitat...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com