Preparation method of sgRNA and HaCaT cell model targeting human genome sequence at high-frequency integration site of high-risk HPV
A technology of integration sites and cell models, applied in the field of medical biology, can solve problems such as cell models without HPV gene integration
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0067] Example 1 Construction of sgRNA targeting plasmid and sgRNA screening
[0068] 1. Construction of sgRNA expression vector
[0069] (1) Design and synthesize sgRNA primers
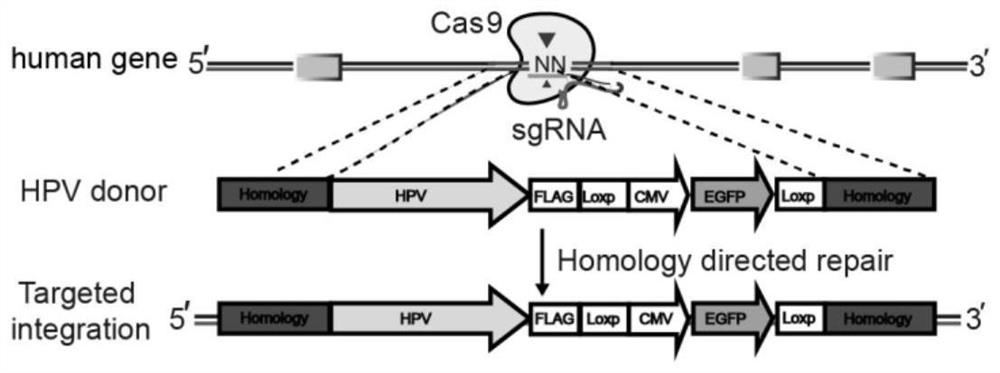
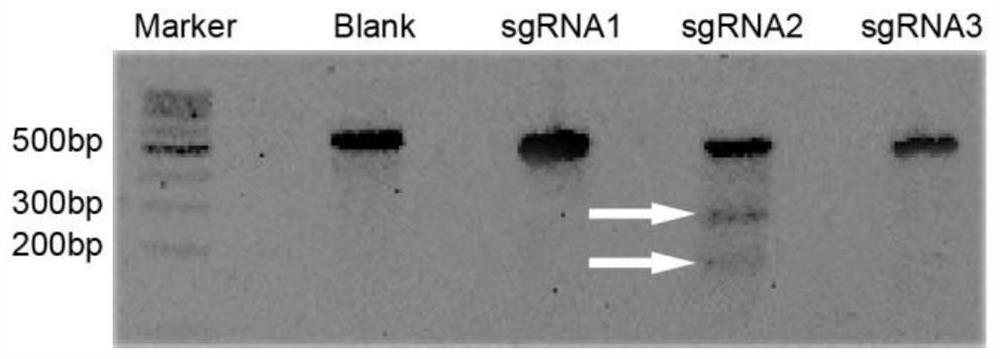
[0070] Select the HPV integration sites with the highest frequency reported in the literature, 13q22 and 8q24. Referring to the HPV18 integration site in the human cervical cancer cell line HeLa, download the 250bp sequence of the 8q24 site 128230508-128230757 region of the human genome from the UCSC (http: / / genome.ucsc.edu / ) website, and use the online software (https : / / zlab.bio / guide-design-resources) to design sgRNA, select the top three with the highest scores, and obtain the sgRNA sequence double-stranded DNA, as shown in Table 1.
[0071]
[0072] Connect CAACCG to the left end of the sense strand of the sgRNA double-stranded DNA sequence, and connect AAACNNNC to both ends of the antisense strand (NNN represents the sequence of the antisense strand of the sgRNA), so as to carry out Bpil d...
Embodiment 2
[0122]Example 2. Preparation of linear Donor fragments containing HPV genes
[0123] (1) The Donor plasmid of the HPV 18 gene and the Donor plasmid of the HPV16 gene were synthesized and constructed by Shanghai Heyuan Biotechnology Co., Ltd. (such as Figure 5-6 shown); wherein, the Donor plasmid containing the high-risk human papillomavirus HPV18 gene (according to the target site of the sgRNA in the 8q24 region, select the upstream and downstream 500bp gene sequences of the CRISPR-Cas9 cleavage site as the left arm of the Donor plasmid respectively 1 and right arm 1, which are connected with the HPV18 gene fragment and the tag gene by a seamless cloning method to construct a Donor plasmid containing the HPV18 gene) consisting of left arm1 (as shown in SEQ ID No.8), HPV18 gene fragment, The tag gene and right arm1 (as shown in SEQ ID No.9) are constructed downstream of the ad element of the Donor backbone plasmid;
[0124] The HPV18 gene fragment is shown in SEQ ID No.7, com...
Embodiment 3
[0136] Example 3 Preparation of HaCaT cell model with site-specific integration of high-risk HPV18 gene
[0137] (1) Co-transfection of sgRNA plasmid and linear Donor fragment
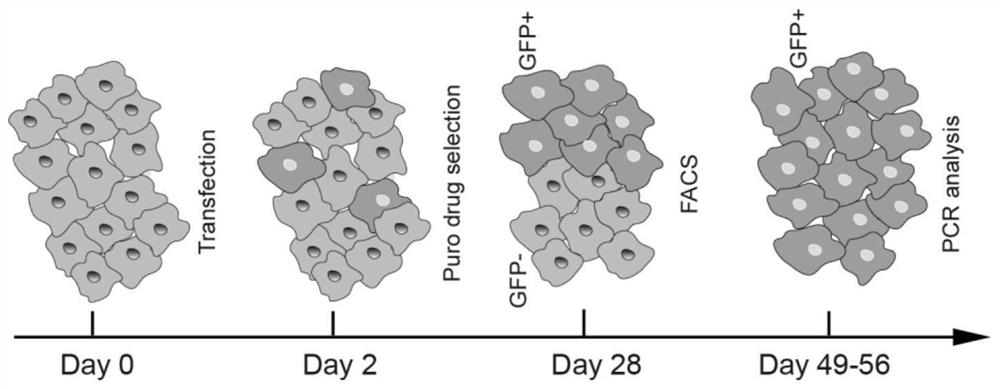
[0138] Use the sgRNA targeting plasmid corresponding to 8q24-sgRNA2 obtained in the example and the linear HPV18 gene Donor fragment obtained in Example 2 to transfect the cells, take the HaCaT cells grown in the logarithmic phase, digest and inoculate them in a 6-well plate, and wait until the degree of cell fusion reaches When transfection is about 80%, each well is transfected (1 μg sgRNA + 1 μg Donor). For specific steps, please refer to the instruction manual.
[0139] (2) Cell drug screening flow sorting
[0140] 24 hours after transfection, according to the expression of the tagged gene, observe the green fluorescence of the cells under a fluorescent microscope to determine the transfection efficiency, then replace the cell culture medium with a complete medium containing 50 μM puromycin, and r...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com