Application of the protein nog1 in the regulation of plant yield and grain number per panicle
A protein and grain number per panicle technology, applied in the biological field, can solve problems such as narrow genetic diversity, similar genetic basis and genotype, and constraints on rice yield potential
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0062] Embodiment 1, the discovery of nog1 gene
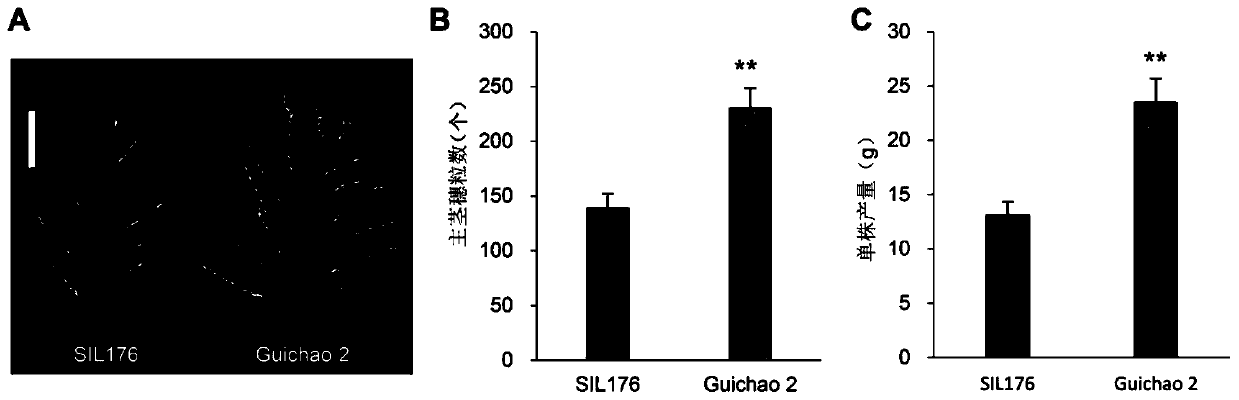
[0063] Using Jiangxi Dongxiang wild rice as the donor parent and Guichao 2 as the recurrent parent, a set of introgressive line populations containing 265 lines (one of which is SIL176) was constructed by crossing and backcrossing. The coverage rate of the wild rice genome in this population is 79.4%, and the yield per plant of 15 lines (one of which is SIL176) is more than 35% lower than that of Guichao 2. The morphology of the main stem panicle, the number of grains on the main stem panicle and the yield per plant of Guichao 2 and SIL176 were compared and counted. The experiment was repeated three times, with 30 plants each time.
[0064] See the experimental results figure 1 (A is the shape of the main stem and ear, bar=5cm; B is the number of grains on the main stem and ear; C is the yield per plant; ** indicates that P<0.01 is extremely significant). The results showed that compared with Guichao 2, the number of grains...
Embodiment 2
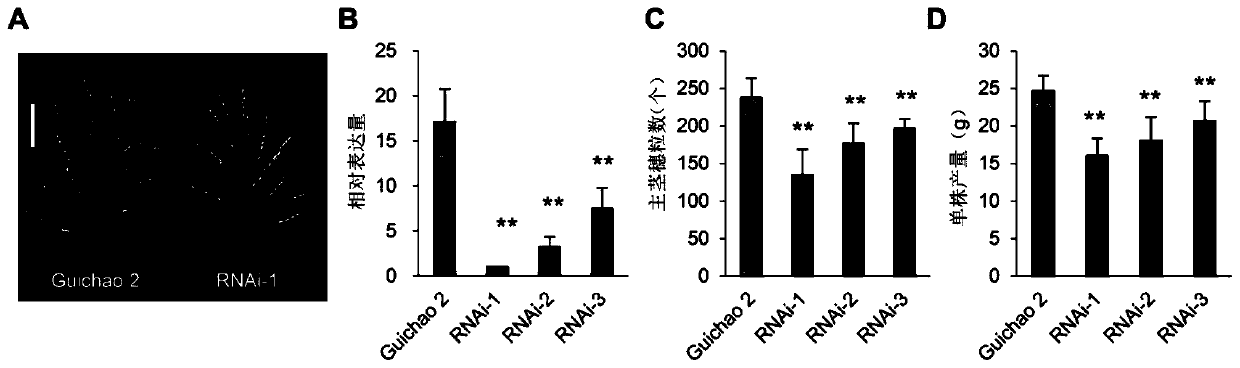
[0066] Example 2, T 2 Obtaining and phenotypic identification of homozygous RNAi interfering strains
[0067] 1. Construction of recombinant plasmid pRNAi-nog1
[0068] The construction steps of the recombinant plasmid pRNAi-nog1 are as follows:
[0069] 1. Synthetic primers
[0070] According to the sequence of the nog1 gene shown in Sequence 1 in the sequence listing, primers 860-rnai-320F, 860-rnai-681R, 860-rnai-681F and 860-rnai-314R were designed and synthesized; the primer sequences are as follows:
[0071] 860-rnai-320F: 5′-GG ACTAGT GGGAGAAAGATGAGGA-3' (the underline is the recognition site of restriction endonuclease SpeI);
[0072] 860-rnai-681R: 5′-TCC GAGCTC GGTCAAAGCCAGGTAC-3' (the underline is the restriction endonuclease SacI recognition site);
[0073] 860-rnai-681F: 5′-CG GGATCC GGTCAAAGCCAGGTAC-3' (the underline is the restriction endonuclease BamHI recognition site);
[0074] 860-rnai-314R: 5′-GG GGTAC CAGAGCTGGGAGAAAGA-3' (the underline is the...
Embodiment 3
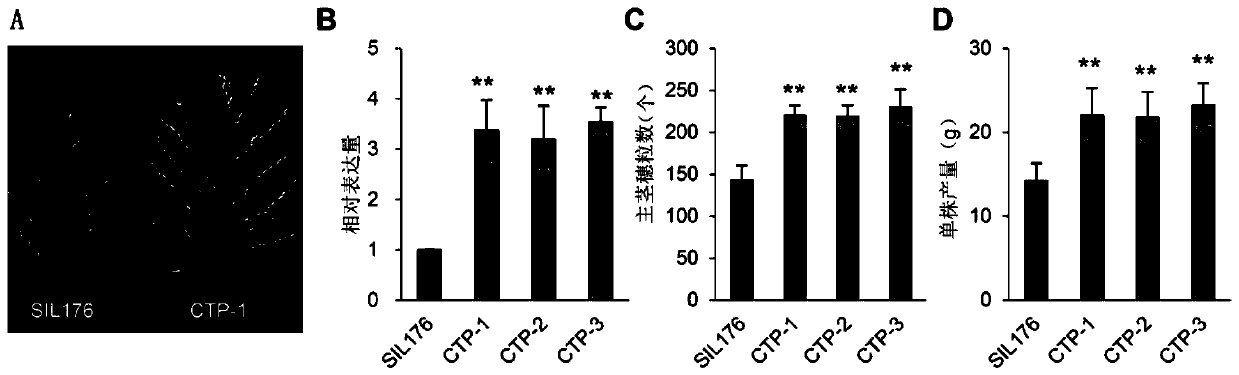
[0103] Example 3, T 2 Generation and phenotypic identification of homozygous complementation lines
[0104] 1. Construction of recombinant plasmid pCAMBIA1300-NOG1
[0105] The construction steps of the recombinant plasmid pCAMBIA1300-NOG1 are as follows:
[0106] 1. Using the 2-week-old seedling experimental material of Guichao 2, extract genomic DNA and use it as a template, use 860HBF: 5'-GA AGATCT CATCTGATGCCTCATACTGA-3' (the underline is the restriction endonuclease BglII recognition site) and 860HBR: 5'-CCG ACGCGT CATGCTTAGGCTGTTGAT-3' (the underline is the recognition site of the restriction endonuclease MluI) was used as a primer for PCR amplification to obtain a PCR amplification product of about 7 kb.
[0107] 2. Digest the PCR amplification product with restriction endonucleases BglII and MluI, and recover the digested product.
[0108] 3. The transformed plant expression vector pCAMBIA1300 was digested with restriction endonucleases BglII and MluI, and about ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com