Circular RNA for diagnosing RSV infection and application thereof
A circular and versatile technology, applied in the field of circular RNA for the diagnosis of RSV infection, can solve the problems of unreported expression changes and achieve high sensitivity and specificity, good specificity, and high sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0027] This embodiment provides a kit for diagnosing RSV infection, the kit includes primers for detecting the expression level of circular RNA whose circBase ID is hsa_circ_0001426, and the primer sequences are such as SEQ ID NO.2 and SEQ ID NO.3 shown.
[0028] The primers are synthesized by conventional techniques in the art, prepared as a mother solution according to the instructions, and ddH can be used to 2 O was diluted to the working concentration.
[0029] The method for detecting the expression level of the circular RNA using the kit comprises the following steps:
[0030] (1) Total RNA extraction: total RNA was extracted by Trizol method.
[0031] (2) Reverse transcription: cDNA was synthesized using the reverse transcription kit PrimeScript RT reagent Kit with gDNA Eraser. The reaction system is 20ul (10ul of the reaction solution in step 1: 5×gDNA Eraser Buffer 2ul, gDNAEraser 1ul, RNA and water total 7ul; PrimeScript RT Enzyme Mix 1ul; RT Primer Mix 1ul; 5×Pri...
Embodiment 2
[0034] In this example, it is confirmed by high-throughput sequencing that the antiviral gene HERC5 produces circular RNA hsa_circ_0001426.
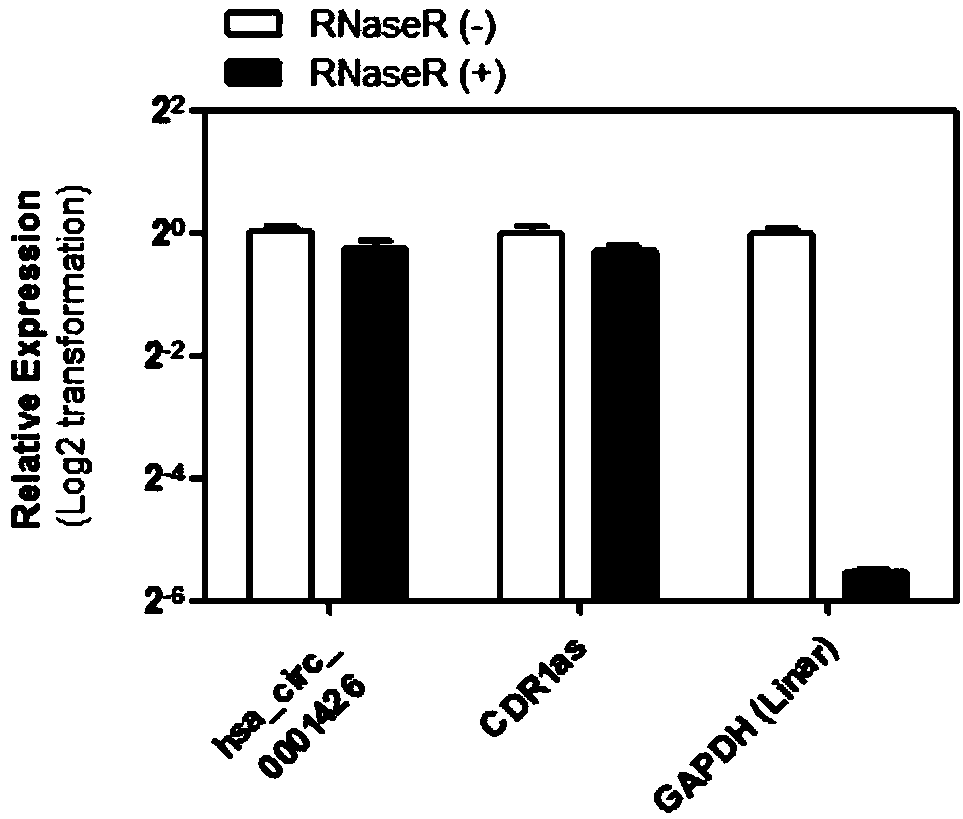
[0035] Methods: Total RNA was extracted from lung cancer cell A549 cells, ribosomal RNA was removed, and then RNase R enzyme was used to degrade the library to further construct the library. The constructed library was sequenced by RNA-Seq using Illumina HiSeq 2500 to analyze the circular RNA related to the antiviral gene HERC5.
[0036] Results: Through analysis, it was found that a circular RNA derived from the HERC5 gene was detected in A549 cells. Its sequence is shown in SEQ ID NO.1.
Embodiment 3
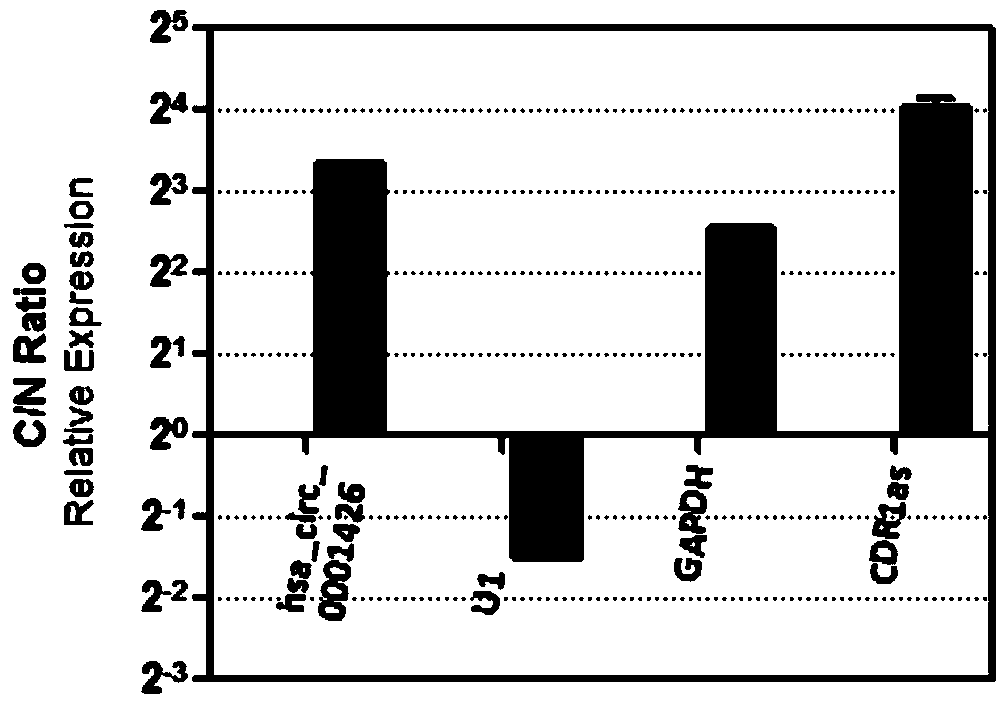
[0038] This example studies the features of hsa_circ_0001426.
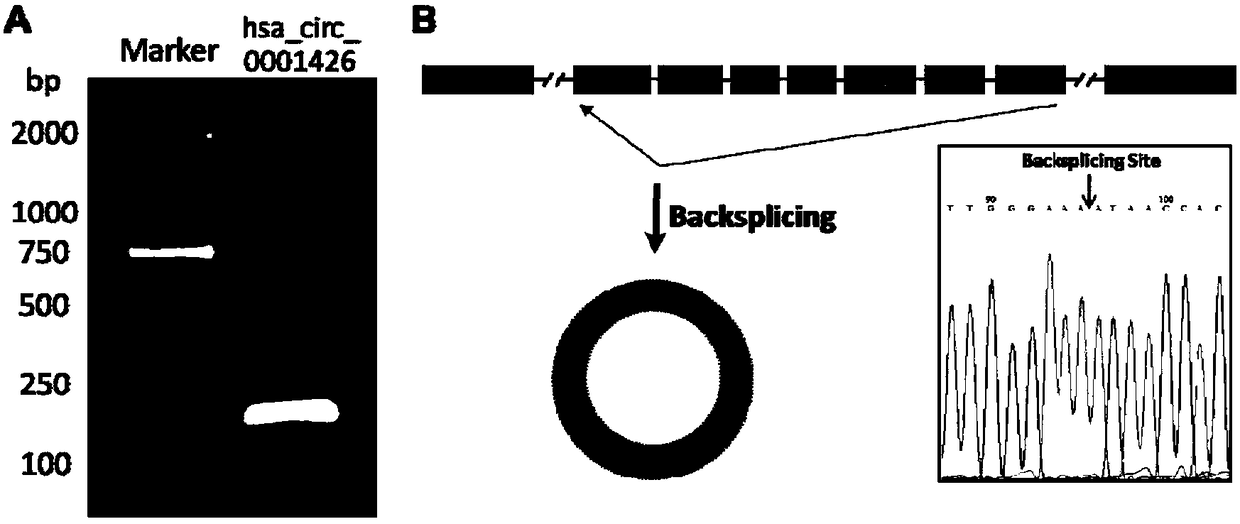
[0039] (1) PCR amplification verification and sequencing verification of the reverse cleavage site of hsa_circ_0001426.
[0040] Method: According to the hsa_circ_0001426 sequence, a pair of bidirectional primers were designed to amplify the sequence containing the reverse cleavage site. The primer pair used included SEQ ID NO.2 and SEQ ID NO.3. Use the cDNA of the cell as a template for PCR amplification, and the reaction system is 20ul (including Takara LA Taq 0.2ul, 10×LA Taq Buffer 2ul, 25mM MgCl 2 2ul, dNTP Mixture 3.2ul, cDNA Template 1ul, forward and reverse primers 1.0ul each, ddH2O 9.6ul); the reaction program was: pre-denaturation at 94°C for 5min; denaturation at 98°C for 10s, annealing at 56°C for 30s, and extension at 72°C for 1min. 34 cycles; 72°C extension for 10 min. The PCR products were detected by 2.0% agarose gel electrophoresis, and sent to Guangzhou Aiji Biotechnology Co., Ltd. for sequenci...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com