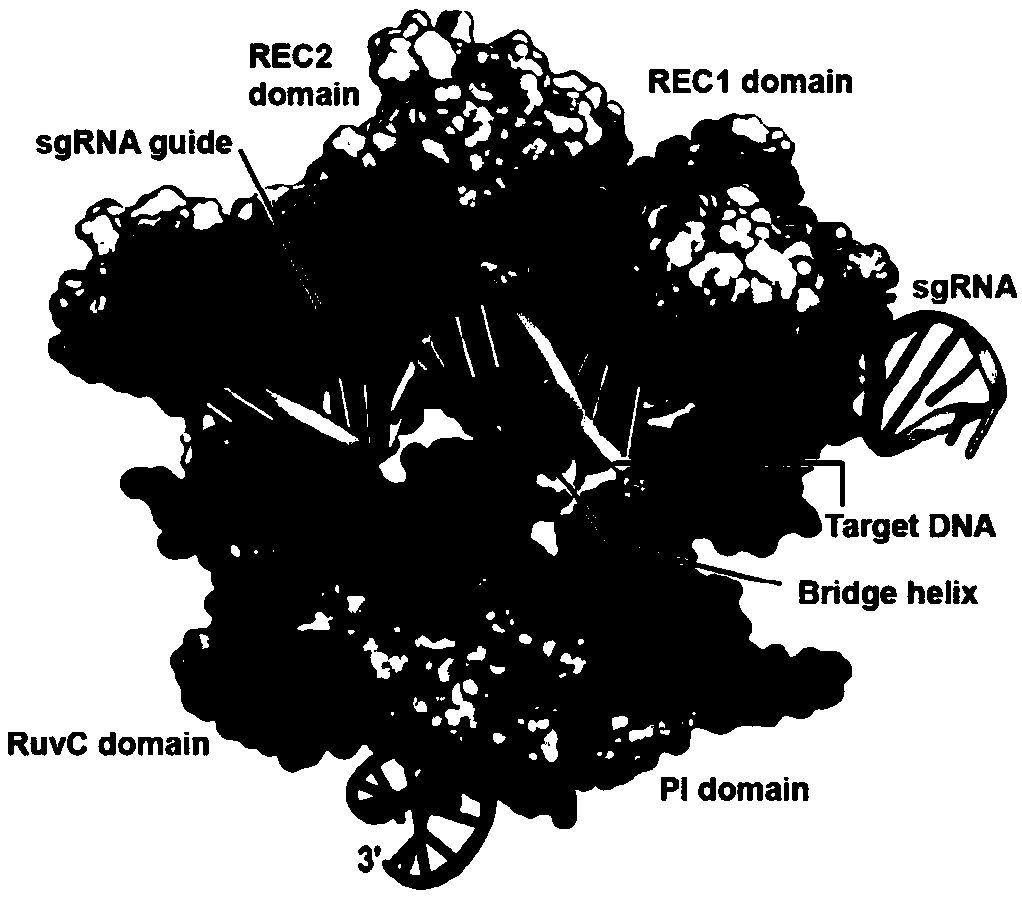
Safe and efficient CRISPR/Cas9 (Clustered Regularly Interspaced Short Palindromic Repeats/CRISPR associated protein 9) gene editing technology
A gene editing and editing technology, applied in the field of gene editing, can solve problems affecting the process of clinical transformation and application, and achieve the effects of strong targeting, simple operation, and high cell transfection efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
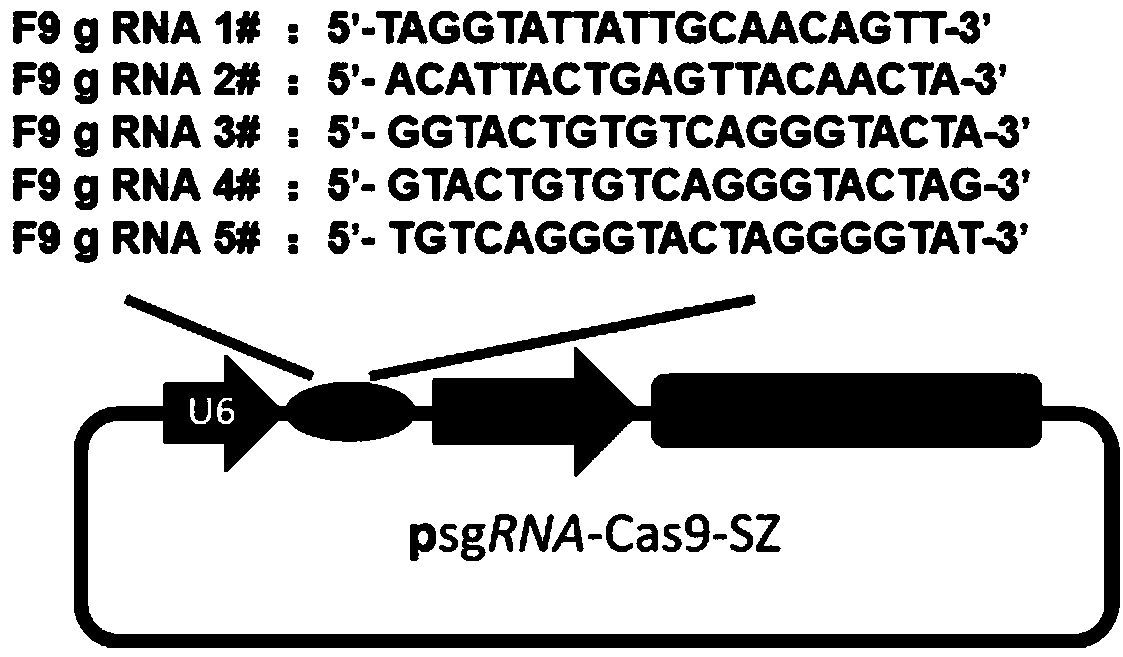
[0023] Hereinafter, the gene editing of HEK293T cells based on the hemophilia IX coagulation factor (F9) gene mutation site is described in detail as a specific gene editing scheme.
[0024] 1. For the F9 gene, design and synthesize 5 sgRNAs, whose sequences are shown in the table below.
[0025]
[0026] Among them, a total of 10 DNA sequences of sgRNA numbers 1-5 can be found in the sequence numbered 2 to 11 in the biological sequence table in the order of the above table, and their names correspond to the names in the above table.
[0027] Each pair of sgRNAs includes two sequences, one of which is the sense strand DNA (S) and the other is the antisense strand DNA (AS). The sequence underlined and in italics is the DNA sequence complementary to the F9 gene.
[0028] 2. First prepare the suspension of sgRNA DNA sense strand and antisense strand so that the final concentration is 100 μM, and anneal according to the following reaction system:
[0029] components ...
Embodiment 2
[0058] Analyze the off-target efficiency of psgRNA-F9-SpCas9-SZ.
[0059] We compared the F9-sgRNA-4 targeting sequence with other sequences in the human genome, and selected the sequence closest to the F9-sgRNA-4 targeting sequence (see Figure 6 sequence shown), specific primers were designed for these sequences, PCR amplification was carried out with genomic DNA as a template, and then the amplified product was sequenced, and the sequencing results (see Figure 7 ) showed that our psgRNA-F9-SpCas9-SZ had no cleavage at these sites.
Embodiment 3
[0061] The target gene was recombined into the F9 gene using psgRNA-F9-SpCas9-SZ and AAV targeting vector DNA, and the recombination efficiency was analyzed.
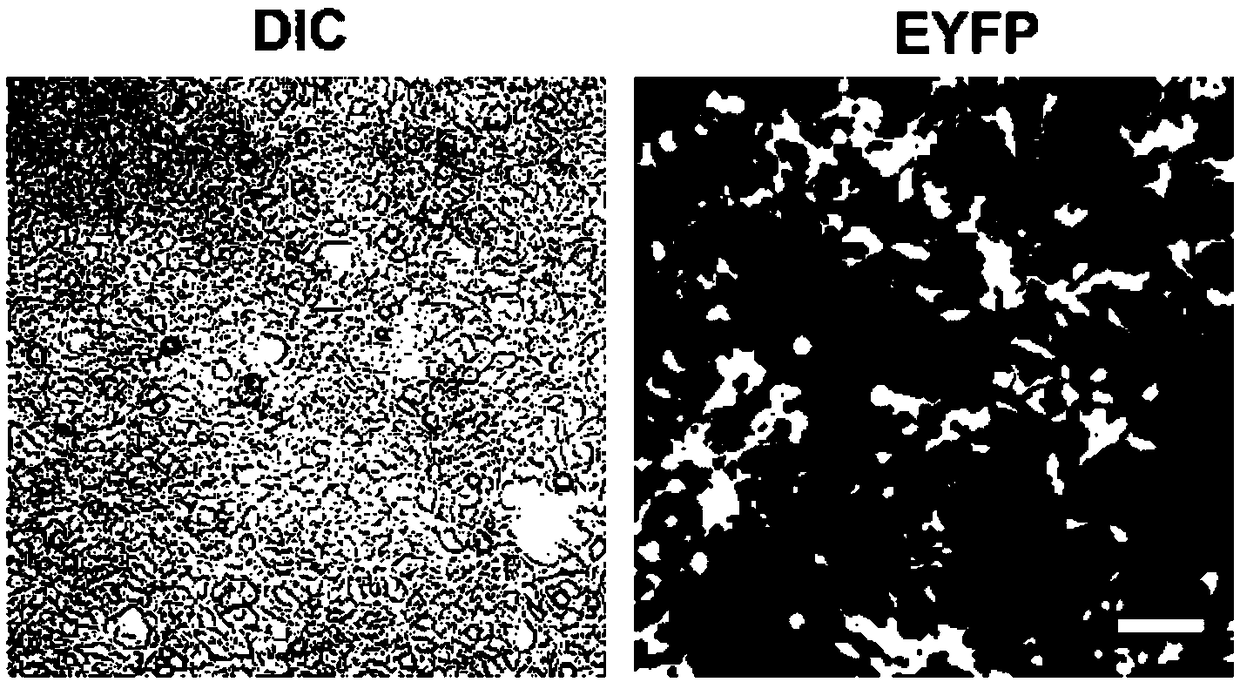
[0062] The pAAV-sgRNA-F9-EF1mini-SpCas9-SZ adeno-associated virus vector and the vector (pAAV-F9-Donor vector) containing the donor gene sequence for repairing the F9 gene were constructed and packaged to infect HEK293T cells. Fluorescence pictures of recombinant cells expressing EGFP are shown in Figure 8 .
[0063] The EGFP-positive cells were sorted by flow cytometry, the genomic DNA of the cells was extracted, and then the sequences between the homologous arms were amplified with specific primers, and the PCR products were sent for sequencing. For sequencing results, see Figure 9 . The results showed that the F9 gene was recombined successfully.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com