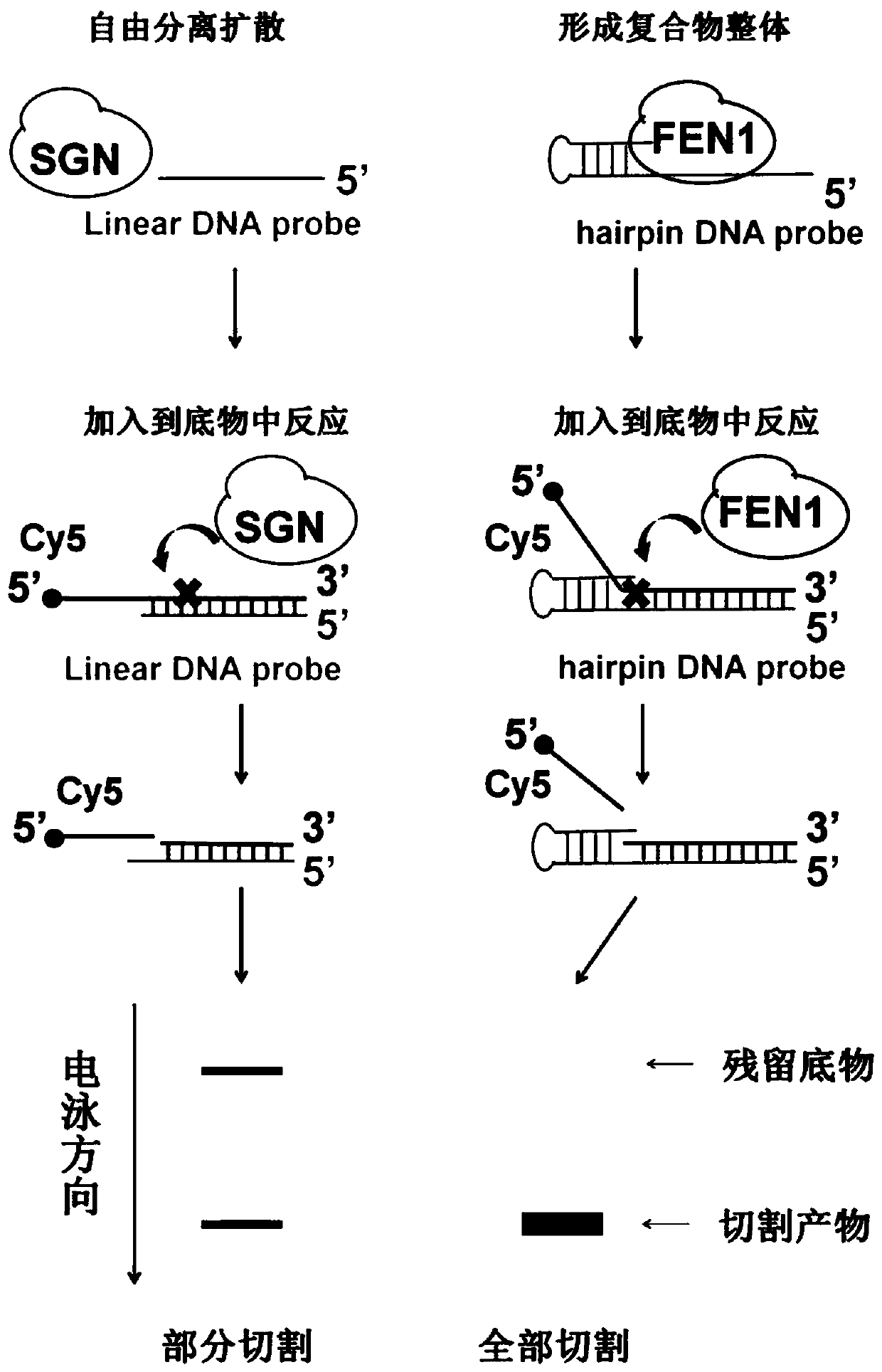
Random nucleic acid editing composition and method
A composition and nucleic acid technology, applied in the field of molecular biology, can solve the problems of poor targeting specificity of Fn1 domain, off-target genomic DNA, low SGN editing efficiency, etc., and achieve the effect of improving gene editing efficiency and strong versatility
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0048] Example 1
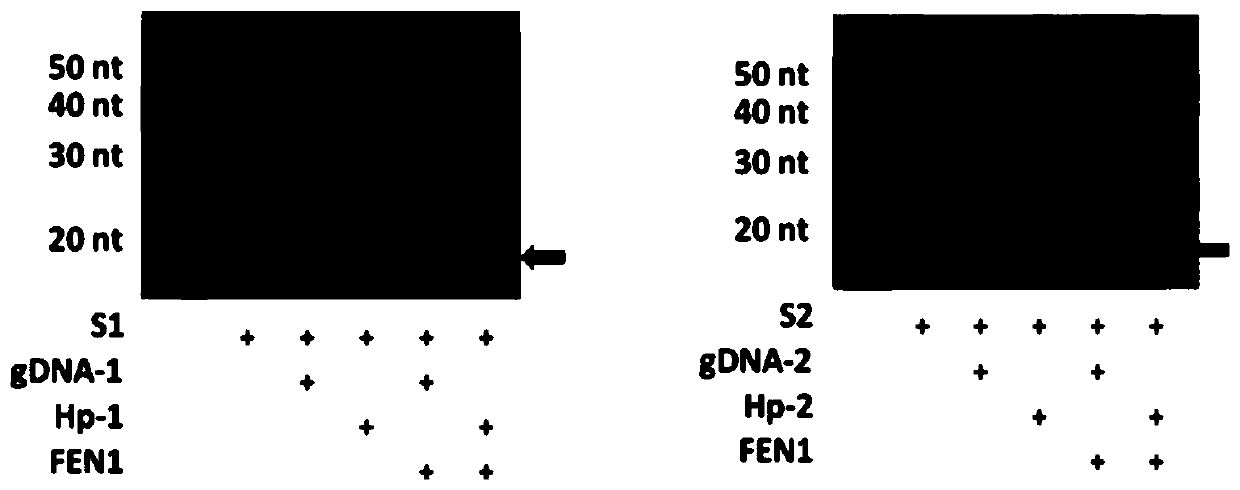
[0049] In order to verify whether AfuFEN1 (the nucleotide sequence is shown in SEQ ID NO.1, and the encoded amino acid sequence is shown in SEQ ID NO.2) can successfully cut target DNA substrates with different sequences, we designed two sequences with completely different DNA substrates S1 and S2, the 5′ end of the target DNA substrate is labeled with fluorescent Cy5, and each substrate matches the corresponding common probe gDNA-1, gDNA-2, and the stem-loop structure hpDNA-1 (referred to as Hp-1) and hpDNA-2 (referred to as Hp-2). The reaction conditions were as follows: 5pmoL hpDNA, 5pmoL target nucleic acid substrate and Mix (10mmol / L MOPS pH 7.5, 0.05% Tween-20, 0.05% Nonidet P40) were thoroughly mixed, and the volume was made up to 10 μL with ultrapure water. Hybridize with the program of "95°C 5min, 55°C 10min", add AfuFEN1 and react at 37°C for 2h. Such as image 3 As shown, when hpDNA and AfuFEN1 exist at the same time, there are obvious cutting...
Embodiment 2
[0051] Embodiment 2
[0052] In order to prove that AfuFEN1 and hpDNA can form a complex independent of the target substrate, the 5' end of Hp-Cy5 was labeled with the fluorescent group Cy5, and incubated with an appropriate amount of AfuFEN1 at 37°C for 0.5h. The mixture was separated by 12% native electrophoresis and imaged by fluorescence. Figure 4 Electrophoretic mobility assay (EMSA) results in demonstrated that AfuFEN1 and Hp-Cy5 can form a complex in the absence of target DNA.
[0053]
Embodiment 3
[0054] Embodiment 3
[0055] We compared the efficiency of SGN+gDNA and AfuFEN1+hpDNA ( Figure 5 -A)). Such as Figure 5 As shown in -B, 1.78 and 0.89 pmol of SGN and gDNA-3 can only cut part of S3, while 0.45, 0.22, 0.11, 0.05 and 0.02 pmol of SGN and gDNA-3 hardly cut S3. At the same time, if Figure 5 As shown in -C, 1.78, 0.89, 0.45, 0.22 pmol of AfuFEN1 and Hp-3 cut S3 almost completely, while 0.11, 0.05 and 0.02 pmol of AfuFEN1 and Hp-3 could cut a part of S3. The results showed that compared with SGN+gDNA, the amount of nuclease required for AfuFEN1+hpDNA to cleave the target protein was reduced by about 40 times ( Figure 5 -D), demonstrating a significant increase in cleavage efficiency.
[0056]
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com